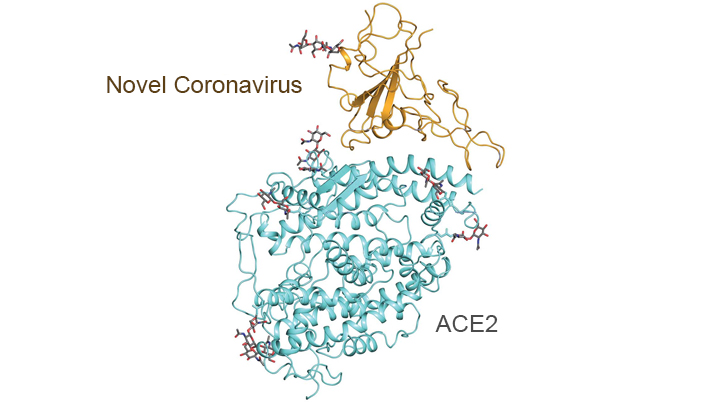
heart disease
All of Us Research Program Participants Fuel Both Scientific and Personal Discovery
Posted on by Josh Denny, M.D., M.S., All of Us Research Program

The NIH’s All of Us Research Program is a historic effort to create an unprecedented research resource that will speed biomedical breakthroughs, transform medicine and advance health equity. To create this resource, we are enrolling at least 1 million people who reflect the diversity of the United States.
At the program’s outset, we promised to make research a two-way street by returning health information to our participant partners. We are now delivering on that promise. We are returning personalized health-related DNA reports to participants who choose to receive them.
That includes me. I signed up to receive my “Medicine and Your DNA” and “Hereditary Disease Risk” reports along with nearly 200,000 other participant partners. I recently read my results, and they hit home, revealing an eye-opening connection between my personal and professional lives.
First, the professional. Before coming to All of Us, I was a practicing physician and researcher at Vanderbilt University, Nashville, TN, where I studied how a person’s genes might affect his or her response to medications. One of the drug-gene interactions that I found most interesting is related to clopidogrel, a drug commonly prescribed to keep arteries open after a major cardiovascular event, like a heart attack, stroke, or placement of a stent.
People with certain gene variations are not able to process this medication well, leaving them in a potentially risky situation. The patient and their health care provider may think the condition is being managed. But, since they can’t process the medication, the patient’s symptoms and risks are likely to increase.
The impact on patients has been seen in numerous studies, including one that I published with colleagues last year in the Journal of Stroke and Cerebrovascular Disease [1]. We found that stroke risk is three times higher in patients who were poor responders to clopidogrel and treated with the drug following a “mini-stroke”—also known as a transient ischemic attack. Other studies have shown that major cardiovascular events were 50 percent more common in individuals who were poor responders to clopidogrel [2]. Importantly, there are alternative therapies that work well for people with this genetic variant.
Now, the personal. Reading my health-related results, I learned that I carry some of these very same gene variations. So, if I ever needed a medicine to manage my risk of blood clots, clopidogrel would not likely work well for me.
Instead, should I ever need treatment, my provider and I could bypass this common first-line therapy and choose an alternate medicine. Getting the right treatment on the first try could cut my chances of a heart attack in half. The benefits of this knowledge don’t stop with me. By choosing to share my findings with family members who may have inherited the same genetic variations, they can discuss it with their health care teams.
Other program participants who choose to receive results will experience the same process of learning more about their health. Nearly all will get actionable information about how their body may process certain medications. A small percentage, 2 to 3 percent, may learn they’re at higher risk of developing several severe hereditary health conditions, such as certain preventable heart diseases and cancers. The program will provide a genetic counselor at no cost to all participants to discuss their results.
To enroll participants who reflect the country’s diverse population, All of Us partners with trusted community organizations around the country. Inclusion is vitally important in the field of genomics research, where available data have long originated mostly from people of European ancestry. In contrast, about 50 percent of the All of Us’ genomic data come from individuals who self-identify with a racial or ethnic minority group.
More than 3,600 research projects are already underway using data contributed by participants from diverse backgrounds. What’s especially exciting about this “ecosystem” of discovery between participants and researchers is that, by contributing their data, participants are helping researchers decode what our DNA is telling us about health across all types of conditions. In turn, those discoveries will deepen what participants can learn.
Those who have stepped up to join All of Us are the heartbeat of this historic research effort to advance personalized approaches in medicine. Their contributions are already fueling new discoveries in numerous areas of health.
At the same time, making good on our promises to our participant partners ensures that the knowledge gained doesn’t only accumulate in a database but is delivered back to participants to help advance their own health journeys. If you’re interested in joining All of Us, we welcome you to learn more.
References:
[1] CYP2C19 loss-of-function is associated with increased risk of ischemic stroke after transient ischemic attack in intracranial atherosclerotic disease. Patel PD, Vimalathas P, Niu X, Shannon CN, Denny JC, Peterson JF, Chitale RV, Fusco MR. J Stroke Cerebrovasc Dis. 2021 Feb;30(2):105464.
[2] Predicting clopidogrel response using DNA samples linked to an electronic health record. Delaney JT, Ramirez AH, Bowton E, Pulley JM, Basford MA, Schildcrout JS, Shi Y, Zink R, Oetjens M, Xu H, Cleator JH, Jahangir E, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Clin Pharmacol Ther. 2012 Feb;91(2):257-263.
Links:
Join All of Us (All of Us/NIH)
NIH’s All of Us Research Program returns genetic health-related results to participants, NIH News Release, December 13, 2022.
NIH’s All of Us Research Program Releases First Genomic Dataset of Nearly 100,000 Whole Genome Sequences, NIH News Release, March 17, 2022.
Funding and Program Partners (All of Us)
Medicine and Your DNA (All of Us)
Clopidogrel Response (National Library of Medicine/NIH)
Hereditary Disease Risk (All of Us)
Preparing for DNA Results: What Is a Genetic Counselor? (All of Us)
Research Projects Directory (All of Us)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes, Centers, and Offices to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 24th in the series of NIH guest posts that will run until a new permanent NIH director is in place.
Millions of Single-Cell Analyses Yield Most Comprehensive Human Cell Atlas Yet
Posted on by Lawrence Tabak, D.D.S., Ph.D.

There are 37 trillion or so cells in our bodies that work together to give us life. But it may surprise you that we still haven’t put a good number on how many distinct cell types there are within those trillions of cells.
That’s why in 2016, a team of researchers from around the globe launched a historic project called the Human Cell Atlas (HCA) consortium to identify and define the hundreds of presumed distinct cell types in our bodies. Knowing where each cell type resides in the body, and which genes each one turns on or off to create its own unique molecular identity, will revolutionize our studies of human biology and medicine across the board.
Since its launch, the HCA has progressed rapidly. In fact, it has already reached an important milestone with the recent publication in the journal Science of four studies that, together, comprise the first multi-tissue drafts of the human cell atlas. This draft, based on analyses of millions of cells, defines more than 500 different cell types in more than 30 human tissues. A second draft, with even finer definition, is already in the works.
Making the HCA possible are recent technological advances in RNA sequencing. RNA sequencing is a topic that’s been mentioned frequently on this blog in a range of research areas, from neuroscience to skin rashes. Researchers use it to detect and analyze all the messenger RNA (mRNA) molecules in a biological sample, in this case individual human cells from a wide range of tissues, organs, and individuals who voluntarily donated their tissues.
By quantifying these RNA messages, researchers can capture the thousands of genes that any given cell actively expresses at any one time. These precise gene expression profiles can be used to catalogue cells from throughout the body and understand the important similarities and differences among them.
In one of the published studies, funded in part by the NIH, a team co-led by Aviv Regev, a founding co-chair of the consortium at the Broad Institute of MIT and Harvard, Cambridge, MA, established a framework for multi-tissue human cell atlases [1]. (Regev is now on leave from the Broad Institute and MIT and has recently moved to Genentech Research and Early Development, South San Francisco, CA.)
Among its many advances, Regev’s team optimized single-cell RNA sequencing for use on cell nuclei isolated from frozen tissue. This technological advance paved the way for single-cell analyses of the vast numbers of samples that are stored in research collections and freezers all around the world.
Using their new pipeline, Regev and team built an atlas including more than 200,000 single-cell RNA sequence profiles from eight tissue types collected from 16 individuals. These samples were archived earlier by NIH’s Genotype-Tissue Expression (GTEx) project. The team’s data revealed unexpected differences among cell types but surprising similarities, too.
For example, they found that genetic profiles seen in muscle cells were also present in connective tissue cells in the lungs. Using novel machine learning approaches to help make sense of their data, they’ve linked the cells in their atlases with thousands of genetic diseases and traits to identify cell types and genetic profiles that may contribute to a wide range of human conditions.
By cross-referencing 6,000 genes previously implicated in causing specific genetic disorders with their single-cell genetic profiles, they identified new cell types that may play unexpected roles. For instance, they found some non-muscle cells that may play a role in muscular dystrophy, a group of conditions in which muscles progressively weaken. More research will be needed to make sense of these fascinating, but vital, discoveries.
The team also compared genes that are more active in specific cell types to genes with previously identified links to more complex conditions. Again, their data surprised them. They identified new cell types that may play a role in conditions such as heart disease and inflammatory bowel disease.
Two of the other papers, one of which was funded in part by NIH, explored the immune system, especially the similarities and differences among immune cells that reside in specific tissues, such as scavenging macrophages [2,3] This is a critical area of study. Most of our understanding of the immune system comes from immune cells that circulate in the bloodstream, not these resident macrophages and other immune cells.
These immune cell atlases, which are still first drafts, already provide an invaluable resource toward designing new treatments to bolster immune responses, such as vaccines and anti-cancer treatments. They also may have implications for understanding what goes wrong in various autoimmune conditions.
Scientists have been working for more than 150 years to characterize the trillions of cells in our bodies. Thanks to this timely effort and its advances in describing and cataloguing cell types, we now have a much better foundation for understanding these fundamental units of the human body.
But the latest data are just the tip of the iceberg, with vast flows of biological information from throughout the human body surely to be released in the years ahead. And while consortium members continue making history, their hard work to date is freely available to the scientific community to explore critical biological questions with far-reaching implications for human health and disease.
References:
[1] Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, Segrè AV, Aguet F, Rozenblatt-Rosen O, Ardlie KG, Regev A, et al. Science. 2022 May 13;376(6594):eabl4290.
[2] Cross-tissue immune cell analysis reveals tissue-specific features in humans. Domínguez Conde C, Xu C, Jarvis LB, Rainbow DB, Farber DL, Saeb-Parsy K, Jones JL,Teichmann SA, et al. Science. 2022 May 13;376(6594):eabl5197.
[3] Mapping the developing human immune system across organs. Suo C, Dann E, Goh I, Jardine L, Marioni JC, Clatworthy MR, Haniffa M, Teichmann SA, et al. Science. 2022 May 12:eabo0510.
Links:
Ribonucleic acid (RNA) (National Human Genome Research Institute/NIH)
Studying Cells (National Institute of General Medical Sciences/NIH)
Regev Lab (Broad Institute of MIT and Harvard, Cambridge, MA)
NIH Support: Common Fund; National Cancer Institute; National Human Genome Research Institute; National Heart, Lung, and Blood Institute; National Institute on Drug Abuse; National Institute of Mental Health; National Institute on Aging; National Institute of Allergy and Infectious Diseases; National Institute of Neurological Disorders and Stroke; National Eye Institute
Decoding Heart-Brain Talk to Prevent Sudden Cardiac Deaths
Posted on by Dr. Francis Collins

As a cardiac electrophysiologist, Deeptankar DeMazumder has worked for years with people at risk for sudden cardiac arrest (SCA). Despite the latest medical advances, less than 10 percent of individuals stricken with an SCA will survive this highly dangerous condition in which irregular heart rhythms, or arrhythmias, cause the heart suddenly to stop beating.
In his role as a physician, DeMazumder keeps a tight focus on the electrical activity in their hearts, doing his best to prevent this potentially fatal event. In his other role, as a scientist at the University of Cincinnati College of Medicine, DeMazumder is also driven by a life-saving aspiration: finding ways to identify at-risk individuals with much greater accuracy than currently possible—and to develop better ways of protecting them from SCAs. He recently received a 2020 NIH Director’s New Innovator Award to pursue one of his promising ideas.
SCAs happen without warning and can cause death within minutes. Poor heart function and abnormal heart rhythms are important risk factors, but it’s not possible today to predict reliably who will have an SCA. However, doctors already routinely capture a wealth of information in electrical signals from the heart using electrocardiograms (ECGs). They also frequently use electroencephalograms (EEGs) to capture electrical activity in the brain.
DeMazumder’s innovative leap is to look at these heart and brain signals jointly, as well as in new ways, during sleep. According to the physician-scientist, sleep is a good time to search for SCA signatures in the electrical crosstalk between the heart and the brain because many other aspects of brain activity quiet down. He also thinks it’s important to pay special attention to what happens to the body’s electrical signals during sleep because most sudden cardiac deaths happen early in the waking hours, for reasons that aren’t well understood.
He has promising preliminary evidence from both animal models and humans suggesting that signatures within heart and brain signals are unique predictors of sudden death, even in people who appear healthy [1]. DeMazumder has already begun developing a set of artificial intelligence algorithms for jointly deciphering waveform signals from the heart, brain, and other body signals [2,3]. These new algorithms associate the waveform signals with a wealth of information available in electronic health records to improve upon the algorithm’s ability to predict catastrophic illness.
DeMazumder credits his curiosity about what he calls the “art and science of healing” to his early childhood experiences and his family’s dedication to community service in India. It taught him to appreciate the human condition, and he has integrated this life-long awareness into his Western medical training and his growing interest in computer science.
For centuries, humans have talked about how true flourishing needs both head and heart. In DeMazumder’s view, science is just beginning to understand the central role of heart-brain conversations in our health. As he continues to capture and interpret these conversations through his NIH-supported work, he hopes to find ways to identify individuals who don’t appear to have serious heart disease but may nevertheless be at high risk for SCA. In the meantime, he will continue to do all he can for the patients in his care.
References:
[1] Mitochondrial ROS drive sudden cardiac death and chronic proteome remodeling in heart failure. Dey S, DeMazumder D, Sidor A, Foster DB, O’Rourke B. Circ Res. 2018;123(3):356-371.
[2] Entropy of cardiac repolarization predicts ventricular arrhythmias and mortality in patients receiving an implantable cardioverter-defibrillator for primary prevention of sudden death. DeMazumder D, Limpitikul WB, Dorante M, et al. Europace. 2016;18(12):1818-1828.
[3] Dynamic analysis of cardiac rhythms for discriminating atrial fibrillation from lethal ventricular arrhythmias. DeMazumder D, Lake DE, Cheng A, et al. Circ Arrhythm Electrophysiol. 2013;6(3):555-561.
Links:
Sudden Cardiac Arrest (National Heart, Lung, and Blood Institute/NIH)
Deeptankar DeMazumder (University of Cincinnati College of Medicine)
DeMazumder Project Information (NIH RePORTER)
NIH Director’s New Innovator Award (Common Fund)
NIH Support: National Heart, Lung, and Blood Institute; Common Fund
All of Us: Partnering Together for the Future of Precision Medicine
Posted on by Dr. Francis Collins

Over the past year, it’s been so inspiring to watch tens of thousands of people across the country selflessly step forward for vaccine trials and other research studies to combat COVID-19. And they are not alone. Many generous folks are volunteering to take part in other types of NIH-funded research that will improve health all across the spectrum, including the more than 360,000 who’ve already enrolled in the pioneering All of Us Research Program.
Now in its second year, All of Us is building a research community of 1 million participant partners to help us learn more about how genetics, environment, and lifestyle interact to influence disease and affect health. So far, more than 80 percent of participants who have completed all the initial enrollment steps are Black, Latino, rural, or from other communities historically underrepresented in biomedical research.
This community will build a diverse foundation for precision medicine, in which care is tailored to the individual, not the average patient as is now often the case. What’s also paradigm shifting about All of Us is its core value of sharing information back with participants about themselves. It is all done responsibly through each participant’s personal All of Us online account and with an emphasis on protecting privacy.
All of Us participants share their health information in many ways, such as taking part in surveys, offering access to their electronic health records, and providing biosamples (blood, urine, and/or saliva). In fact, researchers recently began genotyping and sequencing the DNA in some of those biosamples, and then returning results from analyses to participants who’ve indicated they’d like to receive such information. This first phase of genotyping DNA analysis will provide insights into their genetic ancestry and four traits, including bitter taste perception and tolerance for lactose.
Results of a second sequencing phase of DNA analysis will likely be ready in the coming year. These personalized reports will give interested participants information about how their bodies are likely to react to certain medications and about whether they face an increased risk of developing certain health conditions, such as some types of cancer or heart disease. To help participants better understand the results, they can make a phone appointment with a genetic counselor who is affiliated with the program.
This week, I had the pleasure of delivering the keynote address at the All of Us Virtual Face-to-Face. This lively meeting was attended by a consortium of more than 2,000 All of Us senior staff, program leads with participating healthcare provider organizations and federally qualified health centers, All of Us-supported researchers, community partners, and the all-important participant ambassadors.
If you are interested in becoming part of the All of Us community, I welcome you—there’s plenty of time to get involved! To learn more, just go to Join All of Us.
Links:
All of Us Research Program (NIH)
Join All of Us (NIH)
Modeling Hypertrophic Cardiomyopathy in a Dish
Posted on by Dr. Francis Collins
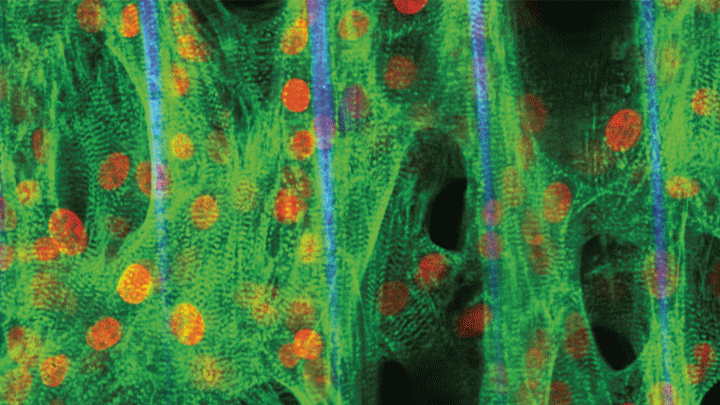
Credit: Zhen Ma, University of California, Berkeley
Researchers have learned in recent years how to grow miniature human hearts in a dish. These “organoids” beat like the real thing and have allowed researchers to model many key aspects of how the heart works. What’s been really tough to model in a dish is how stresses on hearts that are genetically abnormal, such as in inherited familial cardiomyopathies, put people at greater risk for cardiac problems.
Enter the lab-grown human cardiac tissue pictured above. This healthy tissue comprised of the heart’s muscle cells, or cardiomyocytes (green, nuclei in red), was derived from induced pluripotent stem (iPS) cells. These cells are derived from adult skin or blood cells that are genetically reprogrammed to have the potential to develop into many different types of cells, including cardiomyocytes.
An Aspirin a Day for Older People Doesn’t Prolong Healthy Lifespan
Posted on by Dr. Francis Collins

Credit: iStock/thodonal
Many older people who’ve survived a heart attack or stroke take low-dose aspirin every day to help prevent further cardiovascular problems [1]. There is compelling evidence that this works. But should perfectly healthy older folks follow suit?
Most of us would have guessed “yes”—but the answer appears to be “no” when you consider the latest scientific evidence. Recently, a large, international study of older people without a history of cardiovascular disease found that those who took a low-dose aspirin daily over more than 4 years weren’t any healthier than those who didn’t. What’s more, there were some unexpected indications that low-dose aspirin might even boost the risk of death.
Creative Minds: Using Machine Learning to Understand Genome Function
Posted on by Dr. Francis Collins
Science has always fascinated Anshul Kundaje, whether it was biology, physics, or chemistry. When he left his home country of India to pursue graduate studies in electrical engineering at Columbia University, New York, his plan was to focus on telecommunications and computer networks. But a course in computational genomics during his first semester showed him he could follow his interest in computing without giving up his love for biology.
Now an assistant professor of genetics and computer science at Stanford University, Palo Alto, CA, Kundaje has received a 2016 NIH Director’s New Innovator Award to explore not just how the human genome sequence encodes function, but also why it functions in the way that it does. Kundaje even envisions a time when it might be possible to use sophisticated computational approaches to predict the genomic basis of many human diseases.
Next Page