connective tissue
Millions of Single-Cell Analyses Yield Most Comprehensive Human Cell Atlas Yet
Posted on by Lawrence Tabak, D.D.S., Ph.D.

There are 37 trillion or so cells in our bodies that work together to give us life. But it may surprise you that we still haven’t put a good number on how many distinct cell types there are within those trillions of cells.
That’s why in 2016, a team of researchers from around the globe launched a historic project called the Human Cell Atlas (HCA) consortium to identify and define the hundreds of presumed distinct cell types in our bodies. Knowing where each cell type resides in the body, and which genes each one turns on or off to create its own unique molecular identity, will revolutionize our studies of human biology and medicine across the board.
Since its launch, the HCA has progressed rapidly. In fact, it has already reached an important milestone with the recent publication in the journal Science of four studies that, together, comprise the first multi-tissue drafts of the human cell atlas. This draft, based on analyses of millions of cells, defines more than 500 different cell types in more than 30 human tissues. A second draft, with even finer definition, is already in the works.
Making the HCA possible are recent technological advances in RNA sequencing. RNA sequencing is a topic that’s been mentioned frequently on this blog in a range of research areas, from neuroscience to skin rashes. Researchers use it to detect and analyze all the messenger RNA (mRNA) molecules in a biological sample, in this case individual human cells from a wide range of tissues, organs, and individuals who voluntarily donated their tissues.
By quantifying these RNA messages, researchers can capture the thousands of genes that any given cell actively expresses at any one time. These precise gene expression profiles can be used to catalogue cells from throughout the body and understand the important similarities and differences among them.
In one of the published studies, funded in part by the NIH, a team co-led by Aviv Regev, a founding co-chair of the consortium at the Broad Institute of MIT and Harvard, Cambridge, MA, established a framework for multi-tissue human cell atlases [1]. (Regev is now on leave from the Broad Institute and MIT and has recently moved to Genentech Research and Early Development, South San Francisco, CA.)
Among its many advances, Regev’s team optimized single-cell RNA sequencing for use on cell nuclei isolated from frozen tissue. This technological advance paved the way for single-cell analyses of the vast numbers of samples that are stored in research collections and freezers all around the world.
Using their new pipeline, Regev and team built an atlas including more than 200,000 single-cell RNA sequence profiles from eight tissue types collected from 16 individuals. These samples were archived earlier by NIH’s Genotype-Tissue Expression (GTEx) project. The team’s data revealed unexpected differences among cell types but surprising similarities, too.
For example, they found that genetic profiles seen in muscle cells were also present in connective tissue cells in the lungs. Using novel machine learning approaches to help make sense of their data, they’ve linked the cells in their atlases with thousands of genetic diseases and traits to identify cell types and genetic profiles that may contribute to a wide range of human conditions.
By cross-referencing 6,000 genes previously implicated in causing specific genetic disorders with their single-cell genetic profiles, they identified new cell types that may play unexpected roles. For instance, they found some non-muscle cells that may play a role in muscular dystrophy, a group of conditions in which muscles progressively weaken. More research will be needed to make sense of these fascinating, but vital, discoveries.
The team also compared genes that are more active in specific cell types to genes with previously identified links to more complex conditions. Again, their data surprised them. They identified new cell types that may play a role in conditions such as heart disease and inflammatory bowel disease.
Two of the other papers, one of which was funded in part by NIH, explored the immune system, especially the similarities and differences among immune cells that reside in specific tissues, such as scavenging macrophages [2,3] This is a critical area of study. Most of our understanding of the immune system comes from immune cells that circulate in the bloodstream, not these resident macrophages and other immune cells.
These immune cell atlases, which are still first drafts, already provide an invaluable resource toward designing new treatments to bolster immune responses, such as vaccines and anti-cancer treatments. They also may have implications for understanding what goes wrong in various autoimmune conditions.
Scientists have been working for more than 150 years to characterize the trillions of cells in our bodies. Thanks to this timely effort and its advances in describing and cataloguing cell types, we now have a much better foundation for understanding these fundamental units of the human body.
But the latest data are just the tip of the iceberg, with vast flows of biological information from throughout the human body surely to be released in the years ahead. And while consortium members continue making history, their hard work to date is freely available to the scientific community to explore critical biological questions with far-reaching implications for human health and disease.
References:
[1] Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, Segrè AV, Aguet F, Rozenblatt-Rosen O, Ardlie KG, Regev A, et al. Science. 2022 May 13;376(6594):eabl4290.
[2] Cross-tissue immune cell analysis reveals tissue-specific features in humans. Domínguez Conde C, Xu C, Jarvis LB, Rainbow DB, Farber DL, Saeb-Parsy K, Jones JL,Teichmann SA, et al. Science. 2022 May 13;376(6594):eabl5197.
[3] Mapping the developing human immune system across organs. Suo C, Dann E, Goh I, Jardine L, Marioni JC, Clatworthy MR, Haniffa M, Teichmann SA, et al. Science. 2022 May 12:eabo0510.
Links:
Ribonucleic acid (RNA) (National Human Genome Research Institute/NIH)
Studying Cells (National Institute of General Medical Sciences/NIH)
Regev Lab (Broad Institute of MIT and Harvard, Cambridge, MA)
NIH Support: Common Fund; National Cancer Institute; National Human Genome Research Institute; National Heart, Lung, and Blood Institute; National Institute on Drug Abuse; National Institute of Mental Health; National Institute on Aging; National Institute of Allergy and Infectious Diseases; National Institute of Neurological Disorders and Stroke; National Eye Institute
How to Heal Skin Without the Scars
Posted on by Dr. Francis Collins

Most of us can point to a few unwanted scars on our bodies. Every scar tells a story, but people are spending billions of dollars each year trying to hide or get rid of them [1]. What if there was a way to get the wounds on our skin to heal without scarring in the first place?
In a recent paper in the journal Science, a team of NIH-supported researchers has taken an important step in this direction. Working with mice, the researchers deciphered some of the key chemical and physical signals that cause certain skin cells to form tough, fibrous scars while healing a wound [2]. They also discovered how to reprogram them with a topical treatment and respond to injuries more like fetal skin cells, which can patch up wounds in full, regrowing hair, glands, and accessory structures of the skin, and all without leaving a mark.
Of course, mice are not humans. Follow-up research is underway to replicate these findings in larger mammals with skin that’s tighter and more akin to ours. But if the preclinical data hold up, the researchers say they can test in future human clinical trials the anti-scarring drug used in the latest study, which has been commercially available for two decades to treat blood vessel disorders in the eye.
The work comes from Michael Longaker, Shamik Mascharak, and colleagues, Stanford Medicine, Palo Alto, CA. But, to be more precise, the work began with a research project that Longaker was given back in 1987, while a post doc in the lab of Michael Harrison, University of California, San Francisco.
Harrison, a surgeon then performing groundbreaking prenatal surgery, noticed that babies born after undergoing surgery in the womb healed from their surgeries without any scarring. He asked his postdoc to find out why, and Longaker has been trying to answer that question and understand scar formation ever since.
Longaker and his Stanford colleague Geoffrey Gurtner suspected that the difference between healing inside and outside the womb had something to do with tension. Inside the womb, the skin of the unborn is bathed in fluid and develops in a soft, tension-free state. Outside the womb, human skin is exposed to continuous environmental stresses and must continuously remodel and grow to remain viable, which creates a high level of skin tension.
Following up on Longaker and Gurtner’s suspicion, Mascharak found in a series of mouse experiments that a particular class of fibroblast, a type of cell in skin and other connective tissues, activates a gene called Engrailed-1 during scar formation [3]. To see if mechanical stress played a role in this process, Mascharak and team grew mouse fibroblast cells on either a soft, stress-free gel or on a stiff plastic dish that produced mechanical strain. Importantly, they also tried growing the fibroblasts on the same strain-inducing plastic, but in the presence of a chemical that blocked the mechanical-strain signal.
Their studies showed that fibroblasts grown on the tension-free gel didn’t activate the scar-associated genetic program, unlike fibroblasts growing on the stress-inducing plastic. With the chemical that blocked the cells’ ability to sense the mechanical strain, Engrailed-1 didn’t get switched on either.
They also showed the opposite. When tension was applied to healing surgical incisions in mice, it led to an increase in the number of those fibroblast cells expressing Engrailed-1 and thicker scars.
The researchers went on to make another critical finding. The mechanical stress of a fresh injury turns on a genetic program that leads to scar formation, and that program gets switched on through another protein called Yes-associated protein (YAP). When they blocked this protein with an existing eye drug called verteporfin, skin healed more slowly but without any hint of a scar.
It’s worth noting that scars aren’t just a cosmetic issue. Scars differ from unwounded skin in many ways. They lack hair follicles, glands that produce oil and sweat, and nerves for sensing pain or pressure. Because the fibers that make up scar tissue run parallel to each other instead of being more intricately interwoven, scars also lack the flexibility and strength of healthy skin.
These new findings therefore suggest it may one day be possible to allow wounds to heal without compromising the integrity of the skin. The findings also may have implications for many other medical afflictions that involve scarring, such as liver and lung fibrosis, burns, scleroderma, and scarring of heart tissue after a heart attack. That’s also quite a testament to sticking with a good postdoc project, wherever it may lead. One day, it may even improve public health!
References:
[1] Human skin wounds: A major and snowballing threat to public health and the economy. Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, Gottrup F, Gurtner GC, Longaker MT. Wound Repair Regen. 2009 Nov-Dec;17(6):763-771.
[2] Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring.
Mascharak S, desJardins-Park HE, Davitt MF, Griffin M, Borrelli MR, Moore AL, Chen K, Duoto B, Chinta M, Foster DS, Shen AH, Januszyk M, Kwon SH, Wernig G, Wan DC, Lorenz HP, Gurtner GC, Longaker MT. Science. 2021 Apr 23;372(6540):eaba2374.
[3] Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, Weissman IL, Longaker MT. Science. 2015 Apr 17;348(6232):aaa2151.
Links:
Skin Health (National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH)
Scleroderma (NIAMS)
Michael Longaker (Stanford Medicine, Palo Alto, CA)
Geoffrey Gurtner (Stanford Medicine)
NIH Support: National Institute of General Medical Sciences; National Institute of Dental and Craniofacial Research
3D Printing a Human Heart Valve
Posted on by Dr. Francis Collins
It is now possible to pull up the design of a guitar on a computer screen and print out its parts on a 3D printer equipped with special metal or plastic “inks.” The same technological ingenuity is also now being applied with bioinks—printable gels containing supportive biomaterials and/or cells—to print out tissue, bone, blood vessels, and, even perhaps one day, viable organs.
While there’s a long way to go until then, a team of researchers has reached an important milestone in bioprinting collagen and other extracellular matrix proteins that undergird every tissue and organ in the body. The researchers have become so adept at it that they now can print biomaterials that mimic the structural, mechanical, and biological properties of real human tissues.
Take a look at the video. It shows a life-size human heart valve that’s been printed with their improved collagen bioink. As fluid passes through the aortic valve in a lab test, its three leaf-like flaps open and close like the real thing. All the while, the soft, flexible valve withstands the intense fluid pressure, which mimics that of blood flowing in and out of a beating heart.
The researchers, led by NIH grantee Adam Feinberg, Carnegie Mellon University, Pittsburgh, PA, did it with their latest version of a 3D bioprinting technique featured on the blog a few years ago. It’s called: Freeform Reversible Embedding of Suspended Hydrogels v.2.0. Or, just FRESH v2.0.
The FRESH system uses a bioink that consists of collagen (or other soft biomaterials) embedded in a thick slurry of gelatin microparticles and water. While a number of technical improvements have been made to FRESH v. 2.0, the big one was getting better at bioprinting collagen.
The secret is to dissolve the collagen bioink in an acid solution. When extruded into a neutral support bath, the change in pH drives the rapid assembly of collagen. The ability to extrude miniscule amounts and move the needle anywhere in 3D space enables them to produce amazingly complex, high-resolution structures, layer by layer. The porous microstructure of the printed collagen also helps for incorporating human cells. When printing is complete, the support bath easily melts away by heating to body temperature.
As described in Science, in addition to the working heart valve, the researchers have printed a small model of a heart ventricle. By combining collagen with cardiac muscle cells, they found they could actually control the organization of muscle tissue within the model heart chamber. The 3D-printed ventricles also showed synchronized muscle contractions, just like you’d expect in a living, beating human heart!
That’s not all. Using MRI images of an adult human heart as a template, the researchers created a complete organ structure including internal valves, large veins, and arteries. Based on the vessels they could see in the MRI, they printed even tinier microvessels and showed that the structure could support blood-like fluid flow.
While the researchers have focused the potential of FRESH v.2.0 printing on a human heart, in principle the technology could be used for many other organ systems. But there are still many challenges to overcome. A major one is the need to generate and incorporate billions of human cells, as would be needed to produce a transplantable human heart or other organ.
Feinberg reports more immediate applications of the technology on the horizon, however. His team is working to apply FRESH v.2.0 for producing child-sized replacement tracheas and precisely printed scaffolds for healing wounded muscle tissue.
Meanwhile, the Feinberg lab generously shares its designs with the scientific community via the NIH 3D Print Exchange. This innovative program is helping to bring more 3D scientific models online and advance the field of bioprinting. So we can expect to read about many more exciting milestones like this one from the Feinberg lab.
Reference:
[1] 3D bioprinting of collagen to rebuild components of the human heart. Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, Feinberg AW. Science. 2019 Aug 2;365(6452):482-487.
Links:
Tissue Engineering and Regenerative Medicine (National Institute of Biomedical Imaging and Bioengineering/NIH)
Regenerative Biomaterials and Therapeutics Group (Carnegie Mellon University, Pittsburgh, PA)
FluidForm (Acton, MA)
3D Bioprinting Open Source Workshops (Carnegie Mellon)
Video: Adam Feinberg on Tissue Engineering to Treat Human Disease (YouTube)
NIH Support: National Heart, Lung, and Blood Institute; Eunice Kennedy Shriver National Institute of Child Health and Human Development; Common Fund
Snapshots of Life: Muscling in on Development
Posted on by Dr. Francis Collins
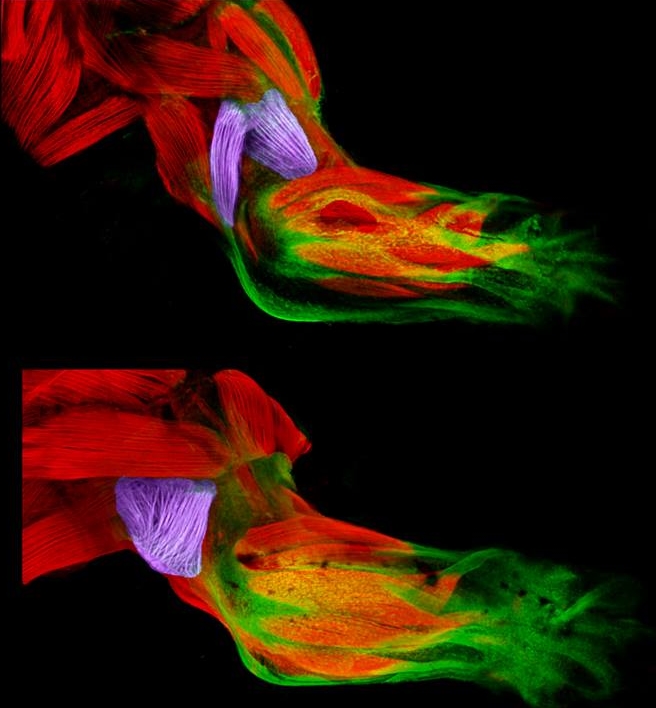
Twice a week, I do an hour of weight training to maintain muscle strength and tone. Millions of Americans do the same, and there’s always a lot of attention paid to those upper arm muscles—the biceps and triceps. Less appreciated is another arm muscle that pumps right along during workouts: the brachialis. This muscle—located under the biceps—helps your elbow flex when you are doing all kinds of things, whether curling a 50-pound barbell or just grabbing a bag of groceries or your luggage out of the car.
Now, scientific studies of the triceps and brachialis are providing important clues about how the body’s 40 different types of limb muscles assume their distinct identities during development [1]. In these images from the NIH-supported lab of Gabrielle Kardon at the University of Utah, Salt Lake City, you see the developing forelimb of a healthy mouse strain (top) compared to that of a mutant mouse strain with a stiff, abnormal gait (bottom).
Snapshots of Life: An Elegant Design
Posted on by Dr. Francis Collins

Credit: David Sleboda and Thomas Roberts, Brown University, Providence, RI
Over the past few years, my blog has highlighted winners from the annual BioArt contest sponsored by the Federation of American Societies for Experimental Biology (FASEB). So, let’s keep a good thing going with one of the amazing scientific images that captured top honors in FASEB’s latest competition: a scanning electron micrograph of the hamstring muscle of a bullfrog.
That’s right, a bullfrog, For decades, researchers have used the American bullfrog, Rana catesbeiana, as a model for studying the physiology and biomechanics of skeletal muscles. My own early work with electron microscopy, as a student at Yale in the 1970s, was devoted to producing images from this very tissue. Thanks to its disproportionately large skeletal muscles, this common amphibian has played a critical role in helping to build the knowledge base for understanding how these muscles work in other organisms, including humans.
Revealed in this picture is the intricate matrix of connective tissue that holds together the frog’s hamstring muscle, with the muscle fibers themselves having been digested away with chemicals. And running diagonally, from lower left to upper right, you can see a band of fibrils made up of a key structural protein called collagen.