precision medicine
All of Us Research Program Participants Fuel Both Scientific and Personal Discovery
Posted on by Josh Denny, M.D., M.S., All of Us Research Program

The NIH’s All of Us Research Program is a historic effort to create an unprecedented research resource that will speed biomedical breakthroughs, transform medicine and advance health equity. To create this resource, we are enrolling at least 1 million people who reflect the diversity of the United States.
At the program’s outset, we promised to make research a two-way street by returning health information to our participant partners. We are now delivering on that promise. We are returning personalized health-related DNA reports to participants who choose to receive them.
That includes me. I signed up to receive my “Medicine and Your DNA” and “Hereditary Disease Risk” reports along with nearly 200,000 other participant partners. I recently read my results, and they hit home, revealing an eye-opening connection between my personal and professional lives.
First, the professional. Before coming to All of Us, I was a practicing physician and researcher at Vanderbilt University, Nashville, TN, where I studied how a person’s genes might affect his or her response to medications. One of the drug-gene interactions that I found most interesting is related to clopidogrel, a drug commonly prescribed to keep arteries open after a major cardiovascular event, like a heart attack, stroke, or placement of a stent.
People with certain gene variations are not able to process this medication well, leaving them in a potentially risky situation. The patient and their health care provider may think the condition is being managed. But, since they can’t process the medication, the patient’s symptoms and risks are likely to increase.
The impact on patients has been seen in numerous studies, including one that I published with colleagues last year in the Journal of Stroke and Cerebrovascular Disease [1]. We found that stroke risk is three times higher in patients who were poor responders to clopidogrel and treated with the drug following a “mini-stroke”—also known as a transient ischemic attack. Other studies have shown that major cardiovascular events were 50 percent more common in individuals who were poor responders to clopidogrel [2]. Importantly, there are alternative therapies that work well for people with this genetic variant.
Now, the personal. Reading my health-related results, I learned that I carry some of these very same gene variations. So, if I ever needed a medicine to manage my risk of blood clots, clopidogrel would not likely work well for me.
Instead, should I ever need treatment, my provider and I could bypass this common first-line therapy and choose an alternate medicine. Getting the right treatment on the first try could cut my chances of a heart attack in half. The benefits of this knowledge don’t stop with me. By choosing to share my findings with family members who may have inherited the same genetic variations, they can discuss it with their health care teams.
Other program participants who choose to receive results will experience the same process of learning more about their health. Nearly all will get actionable information about how their body may process certain medications. A small percentage, 2 to 3 percent, may learn they’re at higher risk of developing several severe hereditary health conditions, such as certain preventable heart diseases and cancers. The program will provide a genetic counselor at no cost to all participants to discuss their results.
To enroll participants who reflect the country’s diverse population, All of Us partners with trusted community organizations around the country. Inclusion is vitally important in the field of genomics research, where available data have long originated mostly from people of European ancestry. In contrast, about 50 percent of the All of Us’ genomic data come from individuals who self-identify with a racial or ethnic minority group.
More than 3,600 research projects are already underway using data contributed by participants from diverse backgrounds. What’s especially exciting about this “ecosystem” of discovery between participants and researchers is that, by contributing their data, participants are helping researchers decode what our DNA is telling us about health across all types of conditions. In turn, those discoveries will deepen what participants can learn.
Those who have stepped up to join All of Us are the heartbeat of this historic research effort to advance personalized approaches in medicine. Their contributions are already fueling new discoveries in numerous areas of health.
At the same time, making good on our promises to our participant partners ensures that the knowledge gained doesn’t only accumulate in a database but is delivered back to participants to help advance their own health journeys. If you’re interested in joining All of Us, we welcome you to learn more.
References:
[1] CYP2C19 loss-of-function is associated with increased risk of ischemic stroke after transient ischemic attack in intracranial atherosclerotic disease. Patel PD, Vimalathas P, Niu X, Shannon CN, Denny JC, Peterson JF, Chitale RV, Fusco MR. J Stroke Cerebrovasc Dis. 2021 Feb;30(2):105464.
[2] Predicting clopidogrel response using DNA samples linked to an electronic health record. Delaney JT, Ramirez AH, Bowton E, Pulley JM, Basford MA, Schildcrout JS, Shi Y, Zink R, Oetjens M, Xu H, Cleator JH, Jahangir E, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Clin Pharmacol Ther. 2012 Feb;91(2):257-263.
Links:
Join All of Us (All of Us/NIH)
NIH’s All of Us Research Program returns genetic health-related results to participants, NIH News Release, December 13, 2022.
NIH’s All of Us Research Program Releases First Genomic Dataset of Nearly 100,000 Whole Genome Sequences, NIH News Release, March 17, 2022.
Funding and Program Partners (All of Us)
Medicine and Your DNA (All of Us)
Clopidogrel Response (National Library of Medicine/NIH)
Hereditary Disease Risk (All of Us)
Preparing for DNA Results: What Is a Genetic Counselor? (All of Us)
Research Projects Directory (All of Us)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes, Centers, and Offices to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 24th in the series of NIH guest posts that will run until a new permanent NIH director is in place.
RADx Initiative: Bioengineering for COVID-19 at Unprecedented Speed and Scale
Posted on by Bruce J. Tromberg, Ph.D., National Institute of Biomedical Imaging and Bioengineering

As COVID-19 rapidly expanded throughout the world in April 2020, many in the biomedical technology community voiced significant concerns about the lack of available diagnostic tests. At that time, testing for SARS-CoV-2, the coronavirus that causes COVID-19, was conducted exclusively in clinical laboratories by order of a health-care provider. “Over the counter” (OTC) tests did not exist, and low complexity point of care (POC) platforms were rare. Fewer than 8 million tests were performed in the U.S. that month, and it was clear that we needed a radical transformation to make tests faster and more accessible.
By February 2022, driven by the Omicron variant surge, U.S. capacity had increased to a new record of more than 1.2 billion tests in a single month. Remarkably, the overwhelming majority of these—more than 85 percent—were “rapid tests” conducted in home and POC settings.
The story behind this practice-changing, “test-at-home” transformation is deeply rooted in technologic and manufacturing innovation. The NIH’s National Institute of Biomedical Imaging and Bioengineering (NIBIB), working collaboratively with multiple partners across NIH, government, academia, and the private sector, has been privileged to play a leading role in this effort via the Rapid Acceleration of Diagnostics (RADx®) initiative. On this two-year anniversary of RADx, we take a brief look back at its formation, impact, and potential for future growth.
On April 24, 2020, Congress recognized that testing was an urgent national need and appropriated $1.5 billion to NIH via an emergency supplement [1]. The goal was to substantially increase the number, type, and availability of diagnostic tests in only five to six months. Since the “normal” commercialization cycle for this type of diagnostic technology is typically more than five years, we needed an entirely new approach . . . fast.
The RADx initiative was launched just five days after that challenging Congressional directive [2]. Four NIH RADx programs were eventually created to support technology development and delivery, with the goal of matching test performance with community needs [3].The first two programs, RADx Tech and RADx Advanced Technology Platforms (ATP), were developed by NIBIB and focused on innovation for rapidly creating, scaling up, and deploying new technologies.
RADx Tech is built around NIBIB’s Point of Care Technologies Research Network (POCTRN) and includes core activities for technology review, test validation, clinical studies, regulatory authorization, and test deployment. Overall, the RADx Tech network includes approximately 900 participants from government, academia, and the private sector with unique capabilities and resources designed to decrease inherent risk and guide technologies from design and development to fully disseminated commercial products.
At the core of RADx Tech operations is the “innovation funnel” rapid review process, popularized as a shark tank [4]. A total of 824 complete applications were submitted during two open calls in a four-month period, beginning April 2020 and during a one-month period in June 2021. Forty-seven projects received phase 1 funding to validate and lower the inherent risk of developing these technologies. Meanwhile, 50 companies received phase 2 contracts to support FDA authorization studies and manufacturing expansion [5]
Beyond test development, RADx Tech has evolved to become a key contributor to the U.S. COVID-19 response. The RADx Independent Test Assessment Program (ITAP) was launched in October 2021 to accelerate regulatory authorization of new tests as a joint effort with the Food and Drug Administration (FDA) [6]. The ITAP acquires analytical and clinical performance data and works closely with FDA and manufacturers to shave weeks to months off the time it normally takes to receive Emergency Use Authorization (EUA).
The RADx Tech program also created a Variant Task Force to monitor the performance of tests against each new coronavirus “variant of concern” that emerges. This helps to ensure that marketed tests continue to remain effective. Other innovative RADx Tech projects include Say Yes! Covid Test, the first online free OTC test distribution program, and Project Rosa, which conducts real-time variant tracking across the country [7].
RADx Tech, by any measure, has exceeded even the most-optimistic expectations. In two years, RADx Tech-supported companies have received 44 EUAs and added approximately 2 billion tests and test products to the U.S. capacity. These remarkable numbers have steadily increased from more than16 million tests in September 2020, just five months after the program was established [8].
RADx Tech has also made significant contributions to the distribution of 1 billion free OTC tests via the government site, COVID.gov/tests. It has also provided critical guidance on serial testing and variants that have improved test performance and changed regulatory practice [9,10]. In addition, the RADx Mobile Application Reporting System (RADx MARS) reduces barriers to test reporting and test-to-treat strategies’ The latter offers immediate treatment options via telehealth or a POC location whenever a positive test result is reported. Finally, the When to Test website provides critical guidance on when and how to test for individuals, groups, and communities.
As we look to the future, RADx Tech has enormous potential to impact the U.S. response to other pathogens, diseases, and future pandemics. Major challenges going forward include improving home tests to work as well as lab platforms and building digital health networks for capturing and reporting test results to public health officials [11].
A recent editorial published in the journal Nature Biotechnology noted, “RADx has spawned a phalanx of diagnostic products to market in just 12 months. Its long-term impact on point of care, at-home, and population testing may be even more profound [12].” We are now poised to advance a new wave of precision medicine that’s led by innovative diagnostic technologies. It represents a unique opportunity to emerge stronger from the pandemic and achieve long-term impact.
References:
[1] Public Law 116 -139—Paycheck Protection Program and Health Care Enhancement Act.
[2] NIH mobilizes national innovation initiative for COVID-19 diagnostics, NIH news release, April 29, 2020.
[3] Rapid scaling up of Covid-19 diagnostic testing in the United States—The NIH RADx Initiative. Tromberg BJ, Schwetz TA, Pérez-Stable EJ, Hodes RJ, Woychik RP, Bright RA, Fleurence RL, Collins FS. N Engl J Med. 2020 Sep 10;383(11):1071-1077.
[4] We need more covid-19 tests. We propose a ‘shark tank’ to get us there. Alexander L. and Blunt R., Washington Post, April 20, 2020.
[5] RADx® Tech/ATP dashboard, National Institute of Biomedical Imaging and Bioengineering, NIH.
[6] New HHS actions add to Biden Administration efforts to increase access to easy-to-use over-the-counter COVID-19 tests. U.S. Department of Health and Human Services Press Office, October 25, 2021.
[7] A method for variant agnostic detection of SARS-CoV-2, rapid monitoring of circulating variants, detection of mutations of biological significance, and early detection of emergent variants such as Omicron. Lai E, et al. medRxiV preprint, January 9, 2022.
[9] Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. Smith RL, et al. J Infect Dis. 2021 Sep 17;224(6):976-982.
[10] Comparison of rapid antigen tests’ performance between Delta (B.1.61.7; AY.X) and Omicron (B.1.1.529; BA1) variants of SARS-CoV-2: Secondary analysis from a serial home self-testing study. Soni A, et al. MedRxiv preprint, March 2, 2022.
[11] Reporting COVID-19 self-test results: The next frontier. Health Affairs, Juluru K., et al. Health Affairs, February 11, 2022.
[12] Radical solutions. Nat Biotechnol. 2021 Apr;39(4):391.
Links:
Get Free At-Home COVID Tests (COVID.gov)
When to Test (Consortia for Improving Medicine with Innovation & Technology, Boston)
RADx Programs (NIH)
RADx® Tech and ATP Programs (National Institute of Biomedical Imaging and Biomedical Engineering/NIH)
Independent Test Assessment Program (NIBIB)
Mobile Application Reporting through Standards (NIBIB)
Point-of-Care Technologies Research Network (POCTRN) (NIBIB)
[Note: Acting NIH Director Lawrence Tabak has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the eighth in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.]
A More Precise Way to Knock Out Skin Rashes
Posted on by Lawrence Tabak, D.D.S., Ph.D.
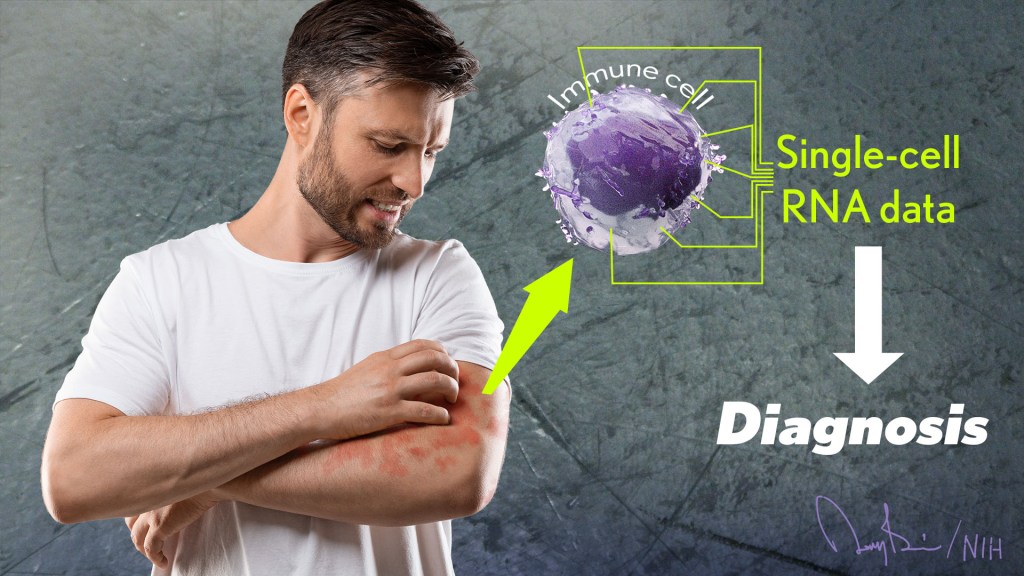
The NIH is committed to building a new era in medicine in which the delivery of health care is tailored specifically to the individual person, not the hypothetical average patient as is now often the case. This new era of “precision medicine” will transform care for many life-threatening diseases, including cancer and chronic kidney disease. But what about non-life-threatening conditions, like the aggravating rash on your skin that just won’t go away?
Recently, researchers published a proof-of-principle paper in the journal Science Immunology demonstrating just how precision medicine for inflammatory skin rashes might work [1]. While more research is needed to build out and further refine the approach, the researchers show it’s now technologically possible to extract immune cells from a patient’s rash, read each cell’s exact inflammatory features, and relatively quickly match them online to the right anti-inflammatory treatment to stop the rash.
The work comes from a NIH-funded team led by Jeffrey Cheng and Raymond Cho, University of California, San Francisco. The researchers focused their attention on two inflammatory skin conditions: atopic dermatitis, the most common type of eczema, which flares up periodically to make skin red and itchy, and psoriasis vulgaris. Psoriasis causes skin cells to build up and form a scaly rash and dry, itchy patches. Together, atopic dermatitis and psoriasis vulgaris affect about 10 percent of U.S. adults.
While the rashes caused by the two conditions can sometimes look similar, they are driven by different sets of immune cells and underlying inflammatory responses. For that reason, distinct biologic therapies, based on antibodies and proteins made from living cells, are now available to target and modify the specific immune pathways underlying each condition.
While biologic therapies represent a major treatment advance for these and other inflammatory conditions, they can miss their targets. Indeed, up to half of patients don’t improve substantially on biologics. Part of the reason for that lack of improvement is because doctors don’t have the tools they need to make firm diagnoses based on what precisely is going on in the skin at the molecular and cellular levels.
To learn more in the new study, the researchers isolated immune cells, focusing primarily on T cells, from the skin of 31 volunteers. They then sequenced the RNA of each cell to provide a telltale portrait of its genomic features. This “single-cell analysis” allowed them to capture high-resolution portraits of 41 different immune cell types found in individual skin samples. That’s important because it offers a much more detailed understanding of changes in the behavior of various immune cells that might have been missed in studies focused on larger groupings of skin cells, representing mixtures of various cell types.
Of the 31 volunteers, seven had atopic dermatitis and eight had psoriasis vulgaris. Three others were diagnosed with other skin conditions, while six had an indeterminate rash with features of both atopic dermatitis and psoriasis vulgaris. Seven others were healthy controls.
The team produced molecular signatures of the immune cells. The researchers then compared the signatures from the hard-to-diagnose rashes to those of confirmed cases of atopic dermatitis and psoriasis. They wanted to see if the signatures could help to reach clearer diagnoses.
The signatures revealed common immunological features as well as underlying differences. Importantly, the researchers found that the signatures allowed them to move forward and classify the indeterminate rashes. The rashes also responded to biologic therapies corresponding to the individuals’ new diagnoses.
Already, the work has identified molecules that help to define major classes of human inflammatory skin diseases. The team has also developed computer tools to help classify rashes in many other cases where the diagnosis is otherwise uncertain.
In fact, the researchers have launched a pioneering website called RashX. It is enabling practicing dermatologists and researchers around the world to submit their single-cell RNA data from their difficult cases. Such analyses are now being done at a small, but growing, number of academic medical centers.
While precision medicine for skin rashes has a long way to go yet before reaching most clinics, the UCSF team is working diligently to ensure its arrival as soon as scientifically possible. Indeed, their new data represent the beginnings of an openly available inflammatory skin disease resource. They ultimately hope to generate a standardized framework to link molecular features to disease prognosis and drug response based on data collected from clinical centers worldwide. It’s a major effort, but one that promises to improve the diagnosis and treatment of many more unusual and long-lasting rashes, both now and into the future.
Reference:
[1] Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Liu Y, Wang H, Taylor M, Cook C, Martínez-Berdeja A, North JP, Harirchian P, Hailer AA, Zhao Z, Ghadially R, Ricardo-Gonzalez RR, Grekin RC, Mauro TM, Kim E, Choi J, Purdom E, Cho RJ, Cheng JB. Sci Immunol. 2022 Apr 15;7(70):eabl9165. {Epub ahead of publication]
Links:
The Promise of Precision Medicine (NIH)
Atopic Dermatitis (National Institute of Arthritis and Musculoskeletal and Skin Diseases /NIH)
Psoriasis (NIAMS/NIH)
RashX (University of California, San Francisco)
Raymond Cho (UCSF)
Jeffrey Cheng (UCSF)
NIH Support: National Institute of Arthritis and Musculoskeletal and Skin Diseases; National Center for Advancing Translational Sciences
All of Us: Release of Nearly 100,000 Whole Genome Sequences Sets Stage for New Discoveries
Posted on by Joshua Denny, M.D., M.S., and Lawrence Tabak, D.D.S., Ph.D.

Nearly four years ago, NIH opened national enrollment for the All of Us Research Program. This historic program is building a vital research community within the United States of at least 1 million participant partners from all backgrounds. Its unifying goal is to advance precision medicine, an emerging form of health care tailored specifically to the individual, not the average patient as is now often the case. As part of this historic effort, many participants have offered DNA samples for whole genome sequencing, which provides information about almost all of an individual’s genetic makeup.
Earlier this month, the All of Us Research Program hit an important milestone. We released the first set of nearly 100,000 whole genome sequences from our participant partners. The sequences are stored in the All of Us Researcher Workbench, a powerful, cloud-based analytics platform that makes these data broadly accessible to registered researchers.
The All of Us Research Program and its many participant partners are leading the way toward more equitable representation in medical research. About half of this new genomic information comes from people who self-identify with a racial or ethnic minority group. That’s extremely important because, until now, over 90 percent of participants in large genomic studies were of European descent. This lack of diversity has had huge impacts—deepening health disparities and hindering scientific discovery from fully benefiting everyone.
The Researcher Workbench also contains information from many of the participants’ electronic health records, Fitbit devices, and survey responses. Another neat feature is that the platform links to data from the U.S. Census Bureau’s American Community Survey to provide more details about the communities where participants live.
This unique and comprehensive combination of data will be key in transforming our understanding of health and disease. For example, given the vast amount of data and diversity in the Researcher Workbench, new diseases are undoubtedly waiting to be uncovered and defined. Many new genetic variants are also waiting to be identified that may better predict disease risk and response to treatment.
To speed up the discovery process, these data are being made available, both widely and wisely. To protect participants’ privacy, the program has removed all direct identifiers from the data and upholds strict requirements for researchers seeking access. Already, more than 1,500 scientists across the United States have gained access to the Researcher Workbench through their institutions after completing training and agreeing to the program’s strict rules for responsible use. Some of these researchers are already making discoveries that promote precision medicine, such as finding ways to predict how to best to prevent vision loss in patients with glaucoma.
Beyond making genomic data available for research, All of Us participants have the opportunity to receive their personal DNA results, at no cost to them. So far, the program has offered genetic ancestry and trait results to more than 100,000 participants. Plans are underway to begin sharing health-related DNA results on hereditary disease risk and medication-gene interactions later this year.
This first release of genomic data is a huge milestone for the program and for health research more broadly, but it’s also just the start. The program’s genome centers continue to generate the genomic data and process about 5,000 additional participant DNA samples every week.
The ultimate goal is to gather health data from at least 1 million or more people living in the United States, and there’s plenty of time to join the effort. Whether you would like to contribute your own DNA and health information, engage in research, or support the All of Us Research Program as a partner, it’s easy to get involved. By taking part in this historic program, you can help to build a better and more equitable future for health research and precision medicine.
Note: Joshua Denny, M.D., M.S., is the Chief Executive Officer of NIH’s All of Us Research Program.
Links:
All of Us Research Program (NIH)
Join All of Us (NIH)
Celebrating NIH Science, Blogs, and Blog Readers!
Posted on by Dr. Francis Collins
Happy holidays to one and all! As you may have heard, this is my last holiday season as the Director of the National Institutes of Health (NIH)—a post that I’ve held for the past 12 years and four months under three U.S. Presidents. And, wow, it really does seem like only yesterday that I started this blog!
At the blog’s outset, I said my goal was to “highlight new discoveries in biology and medicine that I think are game changers, noteworthy, or just plain cool.” More than 1,100 posts, 10 million unique visitors, and 13.7 million views later, I hope you’ll agree that goal has been achieved. I’ve also found blogging to be a whole lot of fun, as well as a great way to expand my own horizons and share a little of what I’ve learned about biomedical advances with people all across the nation and around the world.
So, as I sign off as NIH Director and return to my lab at NIH’s National Human Genome Research Institute (NHGRI), I want to thank everyone who’s ever visited this Blog—from high school students to people with health concerns, from biomedical researchers to policymakers. I hope that the evidence-based information that I’ve provided has helped and informed my readers in some small way.
In this my final post, I’m sharing a short video (see above) that highlights just a few of the blog’s many spectacular images, many of them produced by NIH-funded scientists during the course of their research. In the video, you’ll see a somewhat quirky collection of entries, but hopefully you will sense my enthusiasm for the potential of biomedical research to fight human disease and improve human health—from innovative immunotherapies for treating cancer to the gift of mRNA vaccines to combat a pandemic.
Over the years, I’ve blogged about many of the bold, new frontiers of biomedicine that are now being explored by research teams supported by NIH. Who would have imagined that, within the span of a dozen years, precision medicine would go from being an interesting idea to a driving force behind the largest-ever NIH cohort seeking to individualize the prevention and treatment of common disease? Or that today we’d be deep into investigations of precisely how the human brain works, as well as how human health may benefit from some of the trillions of microbes that call our bodies home?
My posts also delved into some of the amazing technological advances that are enabling breakthroughs across a wide range of scientific fields. These innovative technologies include powerful new ways of mapping the atomic structures of proteins, editing genetic material, and designing improved gene therapies.
So, what’s next for NIH? Let me assure you that NIH is in very steady hands as it heads into a bright horizon brimming with exceptional opportunities for biomedical research. Like you, I look forward to discoveries that will lead us even closer to the life-saving answers that we all want and need.
While we wait for the President to identify a new NIH director, Lawrence Tabak, who has been NIH’s Principal Deputy Director and my right arm for the last decade, will serve as Acting NIH Director. So, keep an eye out for his first post in early January!
As for me, I’ll probably take a little time to catch up on some much-needed sleep, do some reading and writing, and hopefully get out for a few more rides on my Harley with my wife Diane. But there’s plenty of work to do in my lab, where the focus is on type 2 diabetes and a rare disease of premature aging called Hutchinson-Gilford Progeria Syndrome. I’m excited to pursue those research opportunities and see where they lead.
In closing, I’d like to extend my sincere thanks to each of you for your interest in hearing from the NIH Director—and supporting NIH research—over the past 12 years. It’s been an incredible honor to serve you at the helm of this great agency that’s often called the National Institutes of Hope. And now, for one last time, Diane and I take great pleasure in sending you and your loved ones our most heartfelt wishes for Happy Holidays and a Healthy New Year!
Teaching the Immune System to Attack Cancer with Greater Precision
Posted on by Dr. Francis Collins

To protect humans from COVID-19, the Pfizer and Moderna mRNA vaccines program human cells to translate the injected synthetic messenger RNA into the coronavirus spike protein, which then primes the immune system to arm itself against future appearances of that protein. It turns out that the immune system can also be trained to spot and attack distinctive proteins on cancer cells, killing them and leaving healthy cells potentially untouched.
While these precision cancer vaccines remain experimental, researchers continue to make basic discoveries that move the field forward. That includes a recent NIH-funded study in mice that helps to refine the selection of protein targets on tumors as a way to boost the immune response [1]. To enable this boost, the researchers first had to discover a possible solution to a longstanding challenge in developing precision cancer vaccines: T cell exhaustion.
The term refers to the immune system’s complement of T cells and their capacity to learn to recognize foreign proteins, also known as neoantigens, and attack them on cancer cells to shrink tumors. But these responding T cells can exhaust themselves attacking tumors, limiting the immune response and making its benefits short-lived.
In this latest study, published in the journal Cell, Tyler Jacks and Megan Burger, Massachusetts Institute of Technology, Cambridge, help to explain this phenomenon of T cell exhaustion. The researchers found in mice with lung tumors that the immune system initially responds as it should. It produces lots of T cells that target many different cancer-specific proteins.
Yet there’s a problem: various subsets of T cells get in each other’s way. They compete until, eventually, one of those subsets becomes the dominant T cell type. Even when those dominant T cells grow exhausted, they still remain in the tumor and keep out other T cells, which might otherwise attack different neoantigens in the cancer.
Building on this basic discovery, the researchers came up with a strategy for developing cancer vaccines that can “awaken” T cells and reinvigorate the body’s natural cancer-fighting abilities. The strategy might seem counterintuitive. The researchers vaccinated mice with neoantigens that provide a weak but encouraging signal to the immune cells responsible for presenting the distinctive cancer protein target, or antigen, to T cells. It’s those T cells that tend to get suppressed in competition with other T cells.
When the researchers vaccinated the mice with one of those neoantigens, the otherwise suppressed T cells grew in numbers and better targeted the tumor. What’s more, the tumors shrank by more than 25 percent on average.
Research on this new strategy remains in its early stages. The researchers hope to learn if this approach to cancer vaccines might work even better when used in combination with immunotherapy drugs, which unleash the immune system against cancer in other ways.
It’s also possible that the recent and revolutionary success of mRNA vaccines for preventing COVID-19 actually could help. An important advantage of mRNA is that it’s easy for researchers to synthesize once they know the specific nucleic acid sequence of a protein target, and they can even combine different mRNA sequences to make a multiplex vaccine that primes the immune system to recognize multiple neoantigens. Now that we’ve seen how well mRNA vaccines work to prompt a desired immune response against COVID-19, this same technology can be used to speed the development and testing of future vaccines, including those designed precisely to fight cancer.
Reference:
[1] Antigen dominance hierarchies shape TCF1+ progenitor CD8 T cell phenotypes in tumors. Burger ML, Cruz AM, Crossland GE, Gaglia G, Ritch CC, Blatt SE, Bhutkar A, Canner D, Kienka T, Tavana SZ, Barandiaran AL, Garmilla A, Schenkel JM, Hillman M, de Los Rios Kobara I, Li A, Jaeger AM, Hwang WL, Westcott PMK, Manos MP, Holovatska MM, Hodi FS, Regev A, Santagata S, Jacks T. Cell. 2021 Sep 16;184(19):4996-5014.e26.
Links:
Cancer Treatment Vaccines (National Cancer Institute/NIH)
The Jacks Lab (Massachusetts Institute of Technology, Cambridge)
NIH Support: National Cancer Institute; National Heart, Lung, and Blood Institute
A Race-Free Approach to Diagnosing Chronic Kidney Disease
Posted on by Dr. Francis Collins

Race has a long and tortured history in America. Though great strides have been made through the work of leaders like Dr. Martin Luther King, Jr. to build an equal and just society for all, we still have more work to do, as race continues to factor into American life where it shouldn’t. A medical case in point is a common diagnostic tool for chronic kidney disease (CKD), a condition that affects one in seven American adults and causes a gradual weakening of the kidneys that, for some, will lead to renal failure.
The diagnostic tool is a medical algorithm called estimated glomerular filtration rate (eGFR). It involves getting a blood test that measures how well the kidneys filter out a common waste product from the blood and adding in other personal factors to score how well a person’s kidneys are working. Among those factors is whether a person is Black. However, race is a complicated construct that incorporates components that go well beyond biological and genetic factors to social and cultural issues. The concern is that by lumping together Black people, the algorithm lacks diagnostic precision for individuals and could contribute to racial disparities in healthcare delivery—or even runs the risk of reifying race in a way that suggests more biological significance than it deserves.
That’s why I was pleased recently to see the results of two NIH-supported studies published in The New England Journal of Medicine that suggest a way to take race out of the kidney disease equation [1, 2]. The approach involves a new equation that swaps out one blood test for another and doesn’t ask about race.
For a variety of reasons, including socioeconomic issues and access to healthcare, CKD disproportionately affects the Black community. In fact, Blacks with the condition are also almost four times more likely than whites to develop kidney failure. That’s why Blacks with CKD must visit their doctors regularly to monitor their kidney function, and often that visit involves eGFR.
The blood test used in eGFR measures creatinine, a waste product produced from muscle. For about the past 20 years, a few points have been automatically added to the score of African Americans, based on data showing that adults who identify as Black, on average, have a higher baseline level of circulating creatinine. But adjusting the score upward toward normal function runs the risk of making the kidneys seem a bit healthier than they really are and delaying life-preserving dialysis or getting on a transplant list.
A team led by Chi-yuan Hsu, University of California, San Francisco, took a closer look at the current eGFR calculations. The researchers used long-term data from the Chronic Renal Insufficiency Cohort (CRIC) Study, an NIH-supported prospective, observational study of nearly 4,000 racially and ethnically diverse patients with CKD in the U.S. The study design specified that about 40 percent of its participants should identify as Black.
To look for race-free ways to measure kidney function, the researchers randomly selected more than 1,400 of the study’s participants to undergo a procedure that allows kidney function to be measured directly instead of being estimated based on blood tests. The goal was to develop an accurate approach to estimating GFR, the rate of fluid flow through the kidneys, from blood test results that didn’t rely on race.
Their studies showed that simply omitting race from the equation would underestimate GFR in Black study participants. The best solution, they found, was to calculate eGFR based on cystatin C, a small protein that the kidneys filter from the blood, in place of the standard creatinine. Estimation of GFR using cystatin C generated similarly accurate results but without the need to factor in race.
The second NIH-supported study led by Lesley Inker, Tufts Medical Center, Boston, MA, came to similar conclusions. They set out to develop new equations without race using data from several prior studies. They then compared the accuracy of their new eGFR equations to measured GFR in a validation set of 12 other studies, including about 4,000 participants.
Their findings show that currently used equations that include race, sex, and age overestimated measured GFR in Black Americans. However, taking race out of the equation without other adjustments underestimated measured GFR in Black people. Equations including both creatinine and cystatin C, but omitting race, were more accurate. The new equations also led to smaller estimated differences between Black and non-Black study participants.
The hope is that these findings will build momentum toward widespread adoption of cystatin C for estimating GFR. Already, a national task force has recommended immediate implementation of a new diagnostic equation that eliminates race and called for national efforts to increase the routine and timely measurement of cystatin C [3]. This will require a sea change in the standard measurements of blood chemistries in clinical and hospital labs—where creatinine is routinely measured, but cystatin C is not. As these findings are implemented into routine clinical care, let’s hope they’ll reduce health disparities by leading to more accurate and timely diagnosis, supporting the goals of precision health and encouraging treatment of CKD for all people, regardless of their race.
References:
[1] Race, genetic ancestry, and estimating kidney function in CKD. Hsu CY, Yang W, Parikh RV, Anderson AH, Chen TK, Cohen DL, He J, Mohanty MJ, Lash JP, Mills KT, Muiru AN, Parsa A, Saunders MR, Shafi T, Townsend RR, Waikar SS, Wang J, Wolf M, Tan TC, Feldman HI, Go AS; CRIC Study Investigators. N Engl J Med. 2021 Sep 23.
[2] New creatinine- and cystatin C-based equations to estimate GFR without race. Inker LA, Eneanya ND, Coresh J, Tighiouart H, Wang D, Sang Y, Crews DC, Doria A, Estrella MM, Froissart M, Grams ME, Greene T, Grubb A, Gudnason V, Gutiérrez OM, Kalil R, Karger AB, Mauer M, Navis G, Nelson RG, Poggio ED, Rodby R, Rossing P, Rule AD, Selvin E, Seegmiller JC, Shlipak MG, Torres VE, Yang W, Ballew SH,Couture SJ, Powe NR, Levey AS; Chronic Kidney Disease Epidemiology Collaboration. N Engl J Med. 2021 Sep 23.
[3] A unifying approach for GFR estimation: recommendations of the NKF-ASN Task Force on Reassessing the Inclusion of Race in Diagnosing Kidney Disease. Delgado C, Baweja M, Crews DC, Eneanya ND, Gadegbeku CA, Inker LA, Mendu ML, Miller WG, Moxey-Mims MM, Roberts GV, St Peter WL, Warfield C, Powe NR. Am J Kidney Dis. 2021 Sep 22:S0272-6386(21)00828-3.
Links:
Chronic Kidney Disease (National Institute of Diabetes and Digestive and Kidney Diseases/NIH)
Explaining Your Kidney Test Results: A Tool for Clinical Use (NIDDK)
Chronic Renal Insufficiency Cohort Study
Chi-yuan Hsu (University of California, San Francisco)
Lesley Inker (Tufts Medical Center, Boston)
NIH Support: National Institute of Diabetes and Digestive and Kidney Diseases
In Missouri for Grand Opening of Roy Blunt NextGen Precision Health Building
Posted on by Dr. Francis Collins

From Electrical Brain Maps to Learning More About Migraines
Posted on by Dr. Francis Collins

One of life’s greatest mysteries is the brain’s ability to encode something as complex as human behavior. In an effort to begin to unravel this mystery, neuroscientists often zoom in to record the activities of individual neurons. Sometimes they expand their view to look at a specific region of the brain. But if they zoom out farther, neuroscientists can observe many thousands of neurons across the entire brain firing at once to produce electrical oscillations that somehow translate into behaviors as distinct as a smile and a frown. The complexity is truly daunting.
Rainbo Hultman, University of Iowa Carver College of Medicine, Iowa City, realized years ago that by zooming out and finding a way to map all those emergent signals, she could help to change the study of brain function fundamentally. She also realized doing so offered her an opportunity to chip away at cracking the complicated code of the electrical oscillations that translate into such complex behaviors. To pursue her work in this emerging area of “electrical connectomics,” Hultman recently received a 2020 NIH Director’s New Innovator Award to study the most common human neurological disorder: migraine headaches.
A few years ago, Hultman made some impressive progress in electrical connectomics as a post-doctoral researcher in the lab of Kafui Dzirasa at Duke University, Durham, NC. Hultman and her colleagues refined a way to use electrodes to collect electrical field potentials across an unprecedented seven separate mouse brain regions at once. Using machine learning to help make sense of all the data, they uncovered a dynamic, yet reproducible, electrical brain network encoding depression [1].
What’s more, they found that the specific features of this brain-wide network could predict which mice subjected to chronic stress would develop signs of major depressive disorder. As Hultman noted, when measured and mapped in this way, the broad patterns of electrical brain activity, or “Electome factors,” could indicate which mice were vulnerable to stress and which were more resilient.
Moving on to her latest area of research, Hultman is especially intrigued by the fact that people who endure regular migraine attacks often pass through a characteristic sequence of symptoms. These symptoms can include a painful headache on one side of the head; visual disturbances; sensitivity to light, odors, or sound; mood changes; nausea; trouble speaking; and sometimes even paralysis. By studying the broad electrical patterns and networks associated with migraine in mice—simultaneously capturing electrical recordings from 14 brain regions on a millisecond timescale—she wants to understand how brain circuits are linked and work together in ways that produce the complex sequences of migraine symptoms.
More broadly, Hultman wants to understand how migraine and many other disorders affecting the brain lead to a state of heightened sensory sensitivity and how that emerges from integrated neural circuits in the brain. In her studies of migraine, the researcher suspects she might observe some of the same patterns seen earlier in depression. In fact, her team is setting up its experiments to ensure it can identify any brain network features that are shared across important disease states.
By the way, I happen to be one of many people who suffer from migraines, although fortunately not very often in my case. The visual aura of flashing jagged images that starts in the center of my visual field and then gradually moves to the periphery over about 20 minutes is pretty dramatic—a free light show! I’ve wondered what the electrical component of that must be like. But, even with treatment, the headache that follows can be pretty intense.
Hultman also has seen in her own life and family how debilitating migraines can be. Her goal isn’t just to map these neural networks, but to use them to identify where to target future therapeutics. Ultimately, she hopes her work will pave the way for more precise approaches for treating migraine and other brain disorders that are based on the emergent electrical characteristics of each individual’s brain activity. It’s a fascinating proposition, and I certainly look forward to where this research leads and what it may reveal about the fundamentals of how our brains encode complex behaviors and emotions.
Reference:
[1] Brain-wide electrical spatiotemporal dynamics encode depression vulnerability. Hultman R, Ulrich K, Sachs BD, Blount C, Carlson DE, Ndubuizu N, Bagot RC, Parise EM, Vu MT, Gallagher NM, Wang J, Silva AJ, Deisseroth K, Mague SD, Caron MG, Nestler EJ, Carin L, Dzirasa K. Cell. 2018 Mar 22;173(1):166-180.e14.
Links:
Migraine Information Page (National Institute of Neurological Disorders and Stroke/NIH)
Laboratory for Brain-Network Based Molecular Medicine (University of Iowa, Iowa City)
Hultman Project Information (NIH RePORTER)
NIH Director’s New Innovator Award (Common Fund)
NIH Support: Common Fund; National Institute of Mental Health
All of Us: Partnering Together for the Future of Precision Medicine
Posted on by Dr. Francis Collins

Over the past year, it’s been so inspiring to watch tens of thousands of people across the country selflessly step forward for vaccine trials and other research studies to combat COVID-19. And they are not alone. Many generous folks are volunteering to take part in other types of NIH-funded research that will improve health all across the spectrum, including the more than 360,000 who’ve already enrolled in the pioneering All of Us Research Program.
Now in its second year, All of Us is building a research community of 1 million participant partners to help us learn more about how genetics, environment, and lifestyle interact to influence disease and affect health. So far, more than 80 percent of participants who have completed all the initial enrollment steps are Black, Latino, rural, or from other communities historically underrepresented in biomedical research.
This community will build a diverse foundation for precision medicine, in which care is tailored to the individual, not the average patient as is now often the case. What’s also paradigm shifting about All of Us is its core value of sharing information back with participants about themselves. It is all done responsibly through each participant’s personal All of Us online account and with an emphasis on protecting privacy.
All of Us participants share their health information in many ways, such as taking part in surveys, offering access to their electronic health records, and providing biosamples (blood, urine, and/or saliva). In fact, researchers recently began genotyping and sequencing the DNA in some of those biosamples, and then returning results from analyses to participants who’ve indicated they’d like to receive such information. This first phase of genotyping DNA analysis will provide insights into their genetic ancestry and four traits, including bitter taste perception and tolerance for lactose.
Results of a second sequencing phase of DNA analysis will likely be ready in the coming year. These personalized reports will give interested participants information about how their bodies are likely to react to certain medications and about whether they face an increased risk of developing certain health conditions, such as some types of cancer or heart disease. To help participants better understand the results, they can make a phone appointment with a genetic counselor who is affiliated with the program.
This week, I had the pleasure of delivering the keynote address at the All of Us Virtual Face-to-Face. This lively meeting was attended by a consortium of more than 2,000 All of Us senior staff, program leads with participating healthcare provider organizations and federally qualified health centers, All of Us-supported researchers, community partners, and the all-important participant ambassadors.
If you are interested in becoming part of the All of Us community, I welcome you—there’s plenty of time to get involved! To learn more, just go to Join All of Us.
Links:
All of Us Research Program (NIH)
Join All of Us (NIH)
Next Page
