pandemic
Persistence Pays Off: Recognizing Katalin Karikó and Drew Weissman, the 2023 Nobel Prize Winners in Physiology or Medicine
Posted on by Lawrence Tabak, D.D.S., Ph.D.
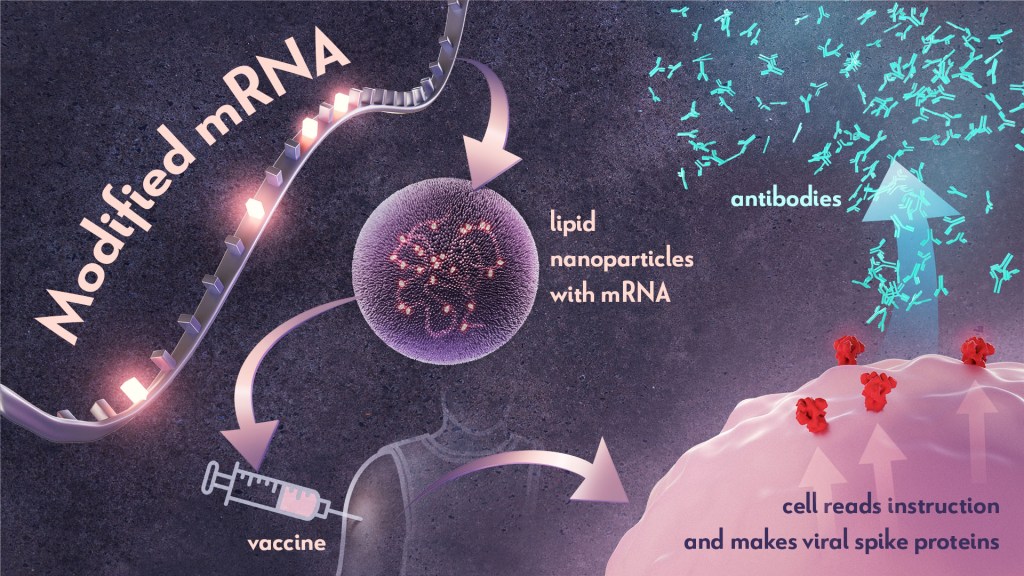
Last week, biochemist Katalin Karikó and immunologist Drew Weissman earned the Nobel Prize in Physiology or Medicine for their discoveries that enabled the development of effective messenger RNA (mRNA) vaccines against COVID-19. On behalf of the NIH community, I’d like to congratulate Karikó and Weissman and thank them for their persistence in pursuing their investigations. NIH is proud to have supported their seminal research, cited by the Nobel Assembly as key publications.1,2,3
While the lifesaving benefits of mRNA vaccines are now clearly realized, Karikó and Weissman’s breakthrough finding in 2005 was not fully appreciated at the time as to why it would be significant. However, their dogged dedication to gaining a better understanding of how RNA interacts with the immune system underscores the often-underappreciated importance of incremental research. Following where the science leads through step-by-step investigations often doesn’t appear to be flashy, but it can end up leading to major advances.
To best describe Karikó and Weissman’s discovery, I’ll first do a quick review of vaccine history. As many of you know, vaccines stimulate our immune systems to protect us from getting infected or from getting very sick from a specific pathogen. Since the late 1700s, scientists have used various approaches to design effective vaccines. Some vaccines introduce a weakened or noninfectious version of a virus to the body, while others present only a small part of the virus, like a protein. The immune system detects the weak or partial virus and develops specialized defenses against it. These defenses work to protect us if we are ever exposed to the real virus.
In the early 1990s, scientists began exploring a different approach to vaccines that involved delivering genetic material, or instructions, so the body’s own cells could make the virus proteins that stimulate an immune response.4,5 Because this approach eliminates the step of growing virus or virus protein in the laboratory—which can be difficult to do in very large quantities and can require a lot of time and money—it had potential, in theory, to be a faster and cheaper way to manufacture vaccines.
Scientists were exploring two types of vaccines as part of this new approach: DNA vaccines and messenger RNA (mRNA) vaccines. DNA vaccines deliver an encoded protein recipe that the cell first copies or transcribes before it starts making protein. For mRNA vaccines, the transcription process is done in the laboratory, and the vaccine delivers the “readable” instructions to the cell for making protein. However, mRNA was not immediately a practical vaccine approach due to several scientific hurdles, including that it caused inflammatory reactions that could be unhealthy for people.
Unfazed by the challenges, Karikó and Weissman spent years pursuing research on RNA and the immune system. They had a brilliant idea that they turned into a significant discovery in 2005 when they proved that inserting subtle chemical modifications to lab-transcribed mRNA eliminated the unwanted inflammatory response.1 In later studies, the pair showed that these chemical modifications also increased protein production.2,3 Both discoveries would be critical to advancing the use of mRNA-based vaccines and therapies.
Earlier theories that mRNA could enable rapid vaccine development turned out to be true. By March 2020, the first clinical trial of an mRNA vaccine for COVID-19 had begun enrolling volunteers, and by December 2020, health care workers were receiving their first shots. This unprecedented timeline was only possible because of Karikó and Weissman’s decades of work, combined with the tireless efforts of many academic, industry and government scientists, including several from the NIH intramural program. Now, researchers are exploring how mRNA could be used in vaccines for other infectious diseases and in cancer vaccines.
As an investigator myself, I’m fascinated by how science continues to build on itself—a process that is done out of the public eye. Luckily every year, the Nobel Prize briefly illuminates for the larger public this long arc of scientific discovery. The Nobel Assembly’s recognition of Karikó and Weissman is a tribute to all scientists who do the painstaking work of trying to understand how things work. Many of the tools we have today to better prevent and treat diseases would not have been possible without the brilliance, tenacity and grit of researchers like Karikó and Weissman.
References:
- K Karikó, et al. Suppression of RNA Recognition by Toll-like Receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity DOI: 10.1016/j.immuni.2005.06.008 (2005).
- K Karikó, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy DOI: 10.1038/mt.2008.200 (2008).
- BR Anderson, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Research DOI: 10.1093/nar/gkq347 (2010).
- DC Tang, et al. Genetic immunization is a simple method for eliciting an immune response. Nature DOI: 10.1038/356152a0 (1992).
- F Martinon, et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. European Journal of Immunology DOI: 10.1002/eji.1830230749 (1993).
NIH Support:
Katalin Karikó: National Heart, Lung, and Blood Institute; National Institute of Neurological Disorders and Stroke
Drew Weissman: National Institute of Allergy and Infectious Diseases; National Institute of Dental and Craniofacial Research; National Heart, Lung, and Blood Institute
Welcome to Response Team Members
Posted on by Lawrence Tabak, D.D.S., Ph.D.

RECOVER: What Clinical Research Comes Next for Helping People with Long COVID

“I connected with RECOVER to be a part of the answers that I was looking for when I was at my worst.” Long COVID patient and RECOVER representative, Nitza Rochez (Bronx, NY)
People, like Nitza Rochez, who are living with Long COVID—the wide-ranging health issues that can follow an infection with SARS-CoV-2, the coronavirus that causes COVID-19—experience disabling symptoms with significant physical, emotional and financial consequences.
The NIH has been engaging and listening to Nitza and others living with Long COVID even before the start of its Researching COVID to Enhance Recovery (RECOVER) Initiative. But now, with the launch of RECOVER, patients and those with affected family or community members have joined researchers, clinicians, and experts in their efforts to unlock the mysteries of Long COVID. All have come together to understand what causes the condition, identify who is most at risk, and determine how to prevent and treat it.
RECOVER is unprecedented in its size and scope as the most-diverse, deeply characterized cohort of Long COVID patients. We’ve enlisted the help of many patient volunteers, who have enrolled in observational studies designed to help researchers learn as much as possible about people who have Long COVID.
Indeed, thousands of research participants are now providing health information and undergoing in-depth medical evaluations and tests, enabling investigators to look for trends. Additionally, studies of millions of electronic medical records are providing insights about those who have received care during the pandemic. More than 40 studies are being conducted to identify the causes of disease, potential biomarkers of Long COVID, and new therapeutic targets.
In all, RECOVER’s research assets are voluminous. They involve invaluable contributions from many people and communities, including research volunteers, research investigators, and clinical specialists. In addition, millions of health records and numerous related tissues and specimens are being analyzed for possible leads.
At the center of it all is the National Community Engagement Group (NCEG). The NCEG is comprised of people living with Long COVID and those representing others living with the condition, and it is truly instrumental to the initiative’s progress in understanding how and why SARS-CoV-2 impacts people in different ways. It’s also helping researchers learn why some people recover while others do not.
So far, we’ve learned that people hospitalized with COVID-19 are twice as likely to have Long COVID than those who were not hospitalized for infection. We’ve also learned that members of racial and ethnic minority groups with Long COVID were more likely to have been hospitalized with COVID-19.
Similarly, disparities in Long COVID exist within those living in areas with particular environmental exposures [1], and those who were already burdened by other diseases and conditions—such as diabetes and chronic pulmonary disease [2]. We’ve also discovered that the certain types of symptoms of Long COVID are consistent among patients regardless of which SARS-CoV-2 variant caused their initial infection. Yet, people infected with the earlier variants have a higher number of symptoms than those infected with more recent variants.
Patient experiences have guided and will continue to guide the study designs and trajectory of RECOVER. Now, fueled by the knowledge that we have gained, RECOVER is preparing to advance to the next phase of discovery—testing interventions in clinical trials to see if they can help people with Long COVID.
To prepare, we are beginning to identify potential clinical trial sites. This important step will help us to find the right places with the right staff and capabilities for enrolling the appropriate patient populations needed to implement the studies. We’ll ensure that the public knows when these upcoming clinical trials are ready to enroll.
Of course, the design of these RECOVER clinical trials will be critical, and insights gained from patients have been key in this process. Results from RECOVER study questionnaires, surveys, and discussions with people experiencing Long COVID identified symptom clusters considered to be the most significant and burdensome to patients. These include sleep disorders, “brain fog” (trouble thinking clearly), exercise intolerance and fatigue, and nervous system dysfunction affecting people’s ability to regulate normal body functions like heart rate and body temperature.
These patient observations have effectively guided the design of the clinical trials that will evaluate whether certain interventions and therapies can help alleviate symptoms that are part of these specific clusters. We’re excited to be advancing toward this phase of the initiative and, again, are very grateful to patient representatives like Nitza, quoted above, for getting us to this phase.
Effective evaluation of those treatments will be important, too. Early in the pandemic, while many clinical trials were launching, most were not large enough or did not have the appropriate objectives to define effective treatments for acute COVID-19. This left clinicians with few clear options when faced with patients needing help.
Learning from this experience, the RECOVER trials will be harmonized to ensure coordinated and efficient evaluation of interventions—in other words, all potential therapies will be using the same protocols platforms and the same data elements. This consistency accelerates our understanding and strengthens the certainty of findings.
Given the widespread and diverse impact that the virus has on the body, it is highly likely that more than one treatment will be needed for each kind of patient experience. Finding solutions for everyone—people of all races, ethnicities, genders, ages, and geographic locations—is paramount.
RECOVER patient representative, Juan Lewis, of San Antonio shared with us, “In April 2020, I was fighting for my life, and today I fight for my quality of life. COVID impacted me physically, mentally, socially, and financially.”
For people like Juan who are experiencing debilitating Long COVID symptoms, we know that finding answers as quickly as possible is critical. As we look ahead to the next 12 months, we’ll continue the studies evaluating the underlying causes, risk factors, and outcomes of Long Covid, and we anticipate significant scientific progress on research leading to Long COVID treatments.
Keep an eye on the RECOVER website for updates on our progress, and published findings.
References:
[1] Identifying environmental risk factors for post-acute sequelae of SARS-CoV-2 infection: An EHR-based cohort study from the recover program. Zhang Y, Hu H, Fokaidis V, V CL, Xu J, Zang C, Xu Z, Wang F, Koropsak M, Bian J, Hall J, Rothman RL, Shenkman EA, Wei WQ, Weiner MG, Carton TW, Kaushal R. Environ Adv. 2023 Apr;11:100352.
[2] Identifying who has long COVID in the USA: a machine learning approach using N3C data. Pfaff ER, Girvin AT, Bennett TD, Bhatia A, Brooks IM, Deer RR, Dekermanjian JP, Jolley SE, Kahn MG, Kostka K, McMurry JA, Moffitt R, Walden A, Chute CG, Haendel MA; N3C Consortium. Lancet Digit Health. 2022 Jul;4(7):e532-e541.
Links:
RECOVER: Researching COVID to Enhance Recovery
Long COVID: Ask NIH Leader about Latest Research (YouTube)
NIH Builds Large Nationwide Study Population of Tens of Thousands to Support Research on Long-Term Effects of COVID-19, NIH News Release, September 15, 2021
Understanding Long-Term COVID-19 Symptoms and Enhancing Recovery, NIH Director’s Blog, October 4, 2022.
NIH RECOVER Research Identifies Potential Long COVID Disparities. NIH News Release, February 16, 2023.
NIH RECOVER Listening Session, June 2021 (NIH Videocast)
NIH RECOVER Listening Session: Understanding Long COVID Across Communities of Color and Those Hardest Hit by COVID, January 21, 2022 (NIH Videocast)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes, Centers, and Offices to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 25th in the series of NIH guest posts that will run until a new permanent NIH director is in place.
Experimental mRNA Vaccine May Protect Against All 20 Influenza Virus Subtypes
Posted on by Lawrence Tabak, D.D.S., Ph.D.

Flu season is now upon us, and protecting yourself and loved ones is still as easy as heading to the nearest pharmacy for your annual flu shot. These vaccines are formulated each year to protect against up to four circulating strains of influenza virus, and they generally do a good job of this. What they can’t do is prevent future outbreaks of more novel flu viruses that occasionally spill over from other species into humans, thereby avoiding a future influenza pandemic.
On this latter and more-challenging front, there’s some encouraging news that was published recently in the journal Science [1]. An NIH-funded team has developed a unique “universal flu vaccine” that, with one seasonal shot, that has the potential to build immune protection against any of the 20 known subtypes of influenza virus and protect against future outbreaks.
While this experimental flu vaccine hasn’t yet been tested in people, the concept has shown great promise in advanced pre-clinical studies. Human clinical trials will hopefully start in the coming year. The researchers don’t expect that this universal flu vaccine will prevent influenza infection altogether. But, like COVID-19 vaccines, the new flu vaccine should help to reduce severe influenza illnesses and deaths when a person does get sick.
So, how does one develop a 20-in-1“multivalent” flu vaccine? It turns out that the key is the same messenger RNA (mRNA) technology that’s enabled two of the safe and effective vaccines against COVID-19, which have been so instrumental in fighting the pandemic. This includes the latest boosters from both Pfizer and Moderna, which now offer updated protection against currently circulating Omicron variants.
While this isn’t the first attempt to develop a universal flu vaccine, past attempts had primarily focused on a limited number of conserved antigens. An antigen is a protein or other substance that produces an immune response. Conserved antigens are those that tend to stay the same over time.
Because conserved antigens will look similar in many different influenza viruses, the hope was that vaccines targeting a small number of them would afford some broad influenza protection. But the focus on a strategy involving few antigens was driven largely by practical limitations. Using traditional methods to produce vaccines by growing flu viruses in eggs and isolating proteins, it simply isn’t feasible to include more than about four targets.
That’s where recent advances in mRNA technology come in. What makes mRNA so nifty for vaccines is that all you need to know is the letters, or sequence, that encodes the genetic material of a virus, including the sequences that get translated into proteins.
A research team led by Scott Hensley, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, recognized that the ease of designing and manufacturing mRNA vaccines opened the door to an alternate approach to developing a universal flu vaccine. Rather than limiting themselves to a few antigens, the researchers could make an all-in-one influenza vaccine, encoding antigens from every known influenza virus subtype.
Influenza vaccines generally target portions of a plentiful protein on the viral surface known as hemagglutinin (H). In earlier work, Hensley’s team, in collaboration with Perelman’s mRNA vaccine pioneer Drew Weissman, showed they could use mRNA technology to produce vaccines with H antigens from single influenza viruses [2, 3]. To protect the fragile mRNA molecules that encode a selected H antigen, researchers deliver them to cells inside well-tolerated microscopic lipid shells, or nanoparticles. The same is true of mRNA COVID-19 vaccines. In their earlier studies, the researchers found that when an mRNA vaccine aimed at one flu virus subtype was given to mice and ferrets in the lab, their cells made the encoded H antigen, eliciting protective antibodies.
In this latest study, they threw antigens from all 20 known flu viruses into the mix. This included H antigens from 18 known types of influenza A and two lineages of influenza B. The goal was to develop a vaccine that could teach the immune system to recognize and respond to any of them.
More study is needed, of course, but early indications are encouraging. The vaccine generated strong and broad antibody responses in animals. Importantly, it worked both in animals with no previous immunity to the flu and in those previously infected with flu viruses. That came as good news because past infections and resulting antibodies sometimes can interfere with the development of new antibodies against related viral subtypes.
In more good news, the researchers found that vaccinated mice and ferrets were protected against severe illness when later challenged with flu viruses. Those viruses included some that were closely matched to antigens in the vaccine, along with some that weren’t.
The findings offer proof-of-principle that mRNA vaccines containing a wide range of antigens can offer broad protection against influenza and likely other viruses as well, including the coronavirus strains responsible for COVID-19. The researchers report that they’re moving toward clinical trials in people, with the goal of beginning an early phase 1 trial in the coming year. The hope is that these developments—driven in part by technological advances and lessons learned over the course of the COVID-19 pandemic—will help to mitigate or perhaps even prevent future pandemics.
References:
[1] A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Arevalo CP, Bolton MJ, Le Sage V, Ye N, Furey C, Muramatsu H, Alameh MG, Pardi N, Drapeau EM, Parkhouse K, Garretson T, Morris JS, Moncla LH, Tam YK, Fan SHY, Lakdawala SS, Weissman D, Hensley SE. Science. 2022 Nov 25;378(6622):899-904.
[2] Nucleoside-modified mRNA vaccination partially overcomes maternal antibody inhibition of de novo immune responses in mice. Willis E, Pardi N, Parkhouse K, Mui BL, Tam YK, Weissman D, Hensley SE. Sci Transl Med. 2020 Jan 8;12(525):eaav5701.
[3] Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Pardi N, Parkhouse K, Kirkpatrick E, McMahon M, Zost SJ, Mui BL, Tam YK, Karikó K, Barbosa CJ, Madden TD, Hope MJ, Krammer F, Hensley SE, Weissman D. Nat Commun. 2018 Aug 22;9(1):3361.
Links:
Understanding Flu Viruses (Centers for Disease Control and Prevention, Atlanta)
COVID Research (NIH)
Decades in the Making: mRNA COVID-19 Vaccines (NIH)
Video: mRNA Flu Vaccines: Preventing the Next Pandemic (Penn Medicine, Philadelphia)
Scott Hensley (Perelman School of Medicine at the University of Pennsylvania, Philadelphia)
Weissman Lab (Perelman School of Medicine)
Video: The Story Behind mRNA COVID Vaccines: Katalin Karikó and Drew Weissman (Penn Medicine, Philadelphia)
NIH Support: National Institute for Allergy and Infectious Diseases
Clinical Center Doctors Testing 3D-Printed Miniature Ventilator
Posted on by James K. Gilman, MD, NIH Clinical Center

Here at the NIH Clinical Center, we are proud to be considered a world-renowned research hospital that provides hope through pioneering clinical research to improve human health. But what you may not know is that our doctors are constantly partnering with public and private sectors to come up with innovative technologies that will help to advance health outcomes.
I’m excited to bring to you a story that is perfect example of the ingenuity of our NIH doctors working with global strategic partners to create potentially life-saving technologies. This story begins during the COVID-19 pandemic with the global shortage of ventilators to help patients breathe. Hospitals had a profound need for inexpensive, easy-to-use, rapidly mass-produced resuscitation devices that could be quickly distributed in areas of critical need.
Through strategic partnerships, our Clinical Center doctors learned about and joined an international group of engineers, physicians, respiratory therapists, and patient advocates using their engineering skills to create a ventilator that was functional, affordable, and intuitive. After several iterations and bench testing, they devised a user-friendly ventilator.
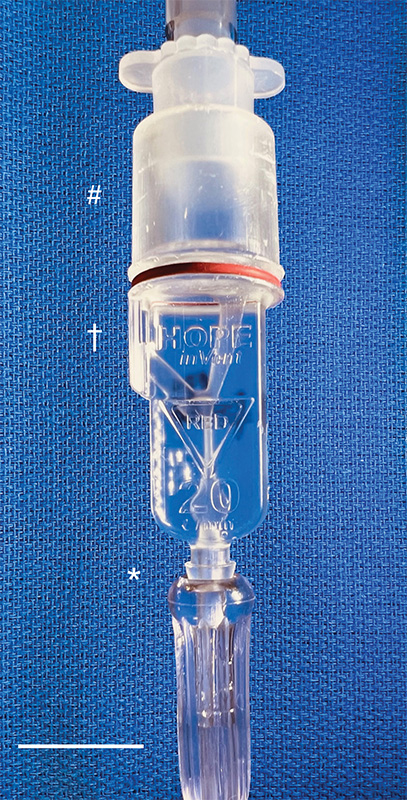
Then, with the assistance of 3D-printing technology, they improved the original design and did something pretty incredible: the team created the smallest single-patient ventilator seen to date. The device is just 2.4 centimeters (about 1 inch) in diameter with a length of 7.4 centimeters (about 3 inches).
A typical ventilator in a hospital obviously is much larger and has a bellows system. It fills with oxygen and then forces it into the lungs followed by the patient passively exhaling. These systems have multiple moving parts, valves, hoses, and electronic or mechanical controls to manage all aspects of the oxygen flow into the lungs.
But our miniature, 3D-printed ventilator is single use, disposable, and has no moving parts. It’s based on principles of fluidics to ventilate patients by automatically oscillating between forced inspiration and assisted expiration as airway pressure changes. It requires only a continuous supply of pressurized oxygen.
The possibilities of this 3D-printed miniature ventilator are broad. The ventilators could be easily used in emergency transport, potentially treating battlefield casualties or responding to disasters and mass casualty events like earthquakes.
While refining a concept is important, the key is converting it to actual use, which our doctors are doing admirably in their preclinical and clinical studies. NIH’s William Pritchard, Andrew Mannes, Brad Wood, John Karanian, Ivane Bakhutashvili, Matthew Starost, David Eckstein, and medical student Sheridan Reed studied and have already tested the ventilators in swine with acute lung injury, a common severe outcome in a number of respiratory threats including COVID-19.
In the study, the doctors tested three versions of the device built to correspond to mild, moderate, and severe lung injury. The respirators provided adequate support for moderate and mild lung injuries, and the doctors recall how amazing it was initially to witness a 190-pound swine ventilated by this miniature ventilator.
The doctors believe that the 3D-printed miniature ventilator is a potential “game changer” from start to finish since it is lifesaving, small, simple to use, can be easily and inexpensively printed and stored, and does not require additional maintenance. They recently published their preclinical trial results in the journal Science Translational Medicine [1].
The NIH team is preparing to initiate first-in-human trials here at the Clinical Center in the coming months. Perhaps, in the not-too-distant future, a device designed to help people breathe could fit into your pocket next to your phone and keys.
Reference:
[1] In-line miniature 3D-printed pressure-cycled ventilator maintains respiratory homeostasis in swine with induced acute pulmonary injury. Pritchard WF, Karanian JW, Jung C, Bakhutashvili I, Reed SL, Starost MF, Froelke BR, Barnes TR, Stevenson D, Mendoza A, Eckstein DJ, Wood BJ, Walsh BK, Mannes AJ. Sci Transl Med. 2022 Oct 12;14(666):eabm8351.
Links:
Clinical Center (NIH)
Andrew Mannes (Clinical Center)
Bradford Wood (Clinical Center)
David Eckstein (Clinical Center)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 21st in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
Gratitude for Biomedical Progress and All Those Who Make It Possible
Posted on by Lawrence Tabak, D.D.S., Ph.D.

It’s good for our health to eat right, exercise, and get plenty of rest. Still, many other things contribute to our sense of wellbeing, including making it a point to practice gratitude whenever we can. With this in mind, I can’t think of a better time than Thanksgiving to recognize just a few of the many reasons that I—and everyone who believes in the mission of the National Institutes of Health (NIH)—have to be grateful.
First, I’m thankful for the many enormously talented people with whom I’ve worked over the past year while performing the duties of the NIH Director. Particular thanks go to those on my immediate team within the Office of the Director. I could not have taken on this challenge without their dedicated support.
I’m also gratified by the continued enthusiasm and support for biomedical research from so many different corners of our society. This includes the many thousands of unsung, patient partners who put their time, effort, and, in some cases, even their lives on the line for the sake of medical progress and promising treatment advances. Without them, clinical research—including the most pivotal clinical trials—simply wouldn’t be possible.
I am most appreciative of the continuing efforts at NIH and across the broader biomedical community to further enable diversity, equity, inclusion, and accessibility within the biomedical research workforce and in all the work that NIH supports.
High on my Thanksgiving list is the widespread availability of COVID-19 bivalent booster shots. These boosters not only guard against older strains of the coronavirus, but also broaden immunity to the newer Omicron variant and its many subvariants. I’m also tremendously grateful for everyone who has—or soon will—get boosted to protect yourself, your loved ones, and your communities as the winter months fast approach.
Another big “thank you” goes out to all the researchers studying Long COVID, the complex and potentially debilitating constellation of symptoms that strikes some people after recovery from COVID-19. I look forward to more answers as this work continues and we certainly couldn’t do it without our patient partners.
I’d also like to express my appreciation for the NIH’s institute and center directors who’ve contributed to the NIH Director’s Blog to showcase NIH’s broad and diverse portfolio of promising research.
Finally, a special thanks to all of you who read this blog. As you gather with family and friends to celebrate this Thanksgiving holiday, I hope the time you spend here gives you a few more reasons to feel grateful and appreciate the importance of NIH in turning scientific discovery into better health for all.
Study Shows Benefits of COVID-19 Vaccines and Boosters
Posted on by Lawrence Tabak, D.D.S., Ph.D.

As colder temperatures settle in and people spend more time gathered indoors, cases of COVID-19 and other respiratory illnesses almost certainly will rise. That’s why, along with scheduling your annual flu shot, it’s now recommended that those age 5 and up should get an updated COVID-19 booster shot [1,2]. Not only will these new boosters guard against the original strain of the coronavirus that started the pandemic, they will heighten your immunity to the Omicron variant and several of the subvariants that continue to circulate in the U.S. with devastating effects.
At last count, about 14.8 million people in the U.S.—including me—have rolled up their sleeves to receive an updated booster shot [3]. It’s a good start, but it also means that most Americans aren’t fully up to date on their COVID-19 vaccines. If you or your loved ones are among them, a new study may provide some needed encouragement to make an appointment at a nearby pharmacy or clinic to get boosted [4].
A team of NIH-supported researchers found a remarkably low incidence of severe COVID-19 illness last fall, winter, and spring among more than 1.6 million veterans who’d been vaccinated and boosted. Severe illness was also quite low in individuals without immune-compromising conditions.
These latest findings, published in the journal JAMA, come from a research group led by Dan Kelly, University of California, San Francisco. He and his team conducted their study drawing on existing health data from the Veterans Health Administration (VA) within a time window of July 2021 and May 2022.
They identified 1.6 million people who’d had a primary-care visit within the last two years and were fully vaccinated for COVID-19, which included receiving a booster shot. Almost three-quarters of those identified were 65 and older. Nearly all were male, and more than 70 percent had another pre-existing health condition that put them at greater risk of becoming seriously ill from a COVID-19 infection.
Over a 24-week follow-up period for each fully vaccinated individual, 125 per 10,000 people had a breakthrough infection. That’s about 1 percent. Just 8.9 in 10,000 fully vaccinated people—less than 0.1 percent—died or were hospitalized from COVID-19 pneumonia. Drilling down deeper into the data:
• Individuals with an immune-compromising condition had a very low rate of hospitalization or death. In this group, 39.6 per 10,000 people had a serious breakthrough infection. That translates to 0.3 percent.
• For people with other preexisting health conditions, including diabetes and heart disease, hospitalization or death totaled 0.07 percent, or 6.7 per 10,000 people.
• For otherwise healthy adults aged 65 and older, the incidence of hospitalization or death was 1.9 per 10,000 people, or 0.02 percent.
• For boosted participants 65 or younger with no high-risk conditions, hospitalization or death came to less than 1 per 10,000 people. That comes to less than 0.01 percent.
It’s worth noting that these results reflect a period when the Delta and Omicron variants were circulating, and available boosters still were based solely on the original variant. Heading into this winter, the hope is that the updated “bivalent” boosters from Pfizer and Moderna will offer even broader protection as this terrible virus continues to evolve.
The Centers for Disease Control and Prevention continues to recommend that everyone stay up to date with their COVID-19 vaccines. That means all adults and kids 5 and older are encouraged to get boosted if it has been at least two months since their last COVID-19 vaccine dose. For older people and those with other health conditions, it’s even more important given their elevated risk for severe illness.
What if you’ve had a COVID-19 infection recently? Getting vaccinated or boosted a few months after you’ve had a COVID-19 infection will offer you even better protection in the future.
So, if you are among the millions of Americans who’ve been vaccinated for COVID-19 but are now due for a booster, don’t delay. Get yourself boosted to protect your own health and the health of your loved ones as the holidays approach.
References:
[1] CDC recommends the first updated COVID-19 booster. Centers for Disease Control and Prevention. September 1, 2022.
[2] CDC expands updated COVID-19 vaccines to include children ages 5 through 11. Centers for Disease Control and Prevention, October 12, 2022.
[3] COVID-19 vaccinations in the United States. Centers for Disease Control and Prevention.
[4] Incidence of severe COVID-19 illness following vaccination and booster with BNT162b2, mRNA-1273, and Ad26.COV2.S vaccines. Kelly JD, Leonard S, Hoggatt KJ, Boscardin WJ, Lum EN, Moss-Vazquez TA, Andino R, Wong JK, Byers A, Bravata DM, Tien PC, Keyhani S. JAMA. 2022 Oct 11;328(14):1427-1437.
Links:
COVID-19 Research (NIH)
Dan Kelly (University of California, San Francisco)
NIH Support: National Institute of Allergy and Infectious Diseases
Suicide Prevention Research in a Rapidly Changing World
Posted on by Joshua A. Gordon, M.D., Ph.D., National Institute of Mental Health

As I sit down to write this blog, the COVID-19 pandemic continues to have a widespread impact, and we’re all trying to figure out our “new normal.” For some, figuring out the new normal has been especially difficult, and that’s something for all of us to consider during September, which is National Suicide Prevention Awareness Month. It’s such an important time to share what we know about suicide prevention and consider how we can further this knowledge to those in need.
At NIH’s National Institute of Mental Health (NIMH), we’ve been asking ourselves: What have we learned about suicide risk and prevention during the pandemic? And how should our research evolve to reflect a rapidly changing world?
Addressing Disparities
Over the last few years, people have been concerned about the pandemic’s impact on suicide rates. So far, data suggest that the overall suicide rate in the U.S. has remained steady. But there is concerning evidence that the pandemic has disproportionately affected suicide risk in historically underserved communities.
For example, data suggest that people in minority racial and ethnic groups experienced greater increases in suicidal thoughts during the pandemic [1]. Additional data indicate that suicide rates may be rising among some young adult racial and ethnic minority groups [2].
Structural racism and other social and environmental factors are major drivers of mental health disparities, and NIMH continues to invest in research to understand how these social determinants of health influence suicide risk. This research includes investigations into the effects of long-term and daily discrimination.
To mitigate these effects, it is critical that we identify specific underlying mechanisms so that we can develop targeted interventions. To this end, NIMH is supporting research in underserved communities to identify suicide risk and the protective factors and effective strategies for reducing this risk (e.g., RFA-MH-22-140, RFA-MH-21-188, RFA-MH-21-187). There are important lessons to be learned that we can’t afford to miss.
Building Solid Foundations
The pandemic also underscored the urgent need to support youth mental health. Indeed, in December 2021, U.S. Surgeon General Dr. Vivek Murthy issued the Advisory on Protecting Youth Mental Health, calling attention to increasing rates of depression and suicidal behaviors among young people. Crucially, the advisory highlighted the need to “recognize that mental health is an essential part of overall health.”
At NIMH, we know that establishing a foundation for good mental health early on can support a person’s overall health and well-being over a lifetime. In light of this, we are investing in research to identify effective prevention efforts that can help set kids on positive mental health trajectories early in life.
Additionally, by re-analyzing research investments already made, we are looking to see whether these early prevention efforts have meaningful impacts on later suicide risk and mental health outcomes. These findings may help to improve a range of systems—such as schools, social services, and health care—to better support kids’ mental health needs.
Improving and Expanding Access
The pandemic has also shown us that telehealth can be an effective means of delivering and increasing access to mental health care. The NIMH has supported research examining telehealth as a tool for improving suicide prevention services, including the use of digital tools that can help extend provider reach and support individuals at risk for suicide.
At the same time, NIMH is investing in work to understand the most effective ways to help providers use evidence-based approaches to prevent suicide. This research helps inform federal partners and others about the best ways to support policies and practices that help prevent suicide deaths.
In July, the Substance Abuse and Mental Health Services Administration (SAMHSA) launched the 988 Suicide & Crisis Lifeline, a three-digit suicide prevention and mental health crisis number. This service builds on the existing National Suicide Prevention Lifeline, allowing anyone to call or text 988 to connect with trained counselors and mental health services. Research supported by NIMH helped build the case for such lifelines, and now we’re calling for research aimed at identifying the best ways to help people use this evolving crisis support system.
Looking Ahead
With these and many other efforts, we are hopeful that people who are at risk for suicidal thoughts and behaviors will be able to access the evidence-based support and services they need. This National Suicide Prevention Awareness Month, I’d like to issue a call to action: Help raise awareness by sharing resources on how to recognize the warning signs for suicide and how to get help. By working together, we can prevent suicide and save lives.
References:
[1] Racial and ethnic disparities in the prevalence of stress and worry, mental health conditions, and increased substance use among adults during the COVID-19 pandemic – United States, April and May 2020. McKnight-Eily LR, Okoro CA, Strine TW, Verlenden J, Hollis ND, Njai R, Mitchell EW, Board A, Puddy R, Thomas C. MMWR Morb Mortal Wkly Rep. 2021 Feb 5;70(5):162-166.
[2] One Year In: COVID-19 and Mental Health. National Institute of Mental Health Director’s Message. April 9, 2021.
Links:
988 Suicide & Crisis Lifeline (Substance Abuse and Mental Health Services Administration, Rockville, MD)
Substance Abuse and Mental Health Services Administration Treatment Locator (SAMHSA)
Help for Mental Illnesses (National Institute of Mental Health/NIH)
Suicide Prevention (NIMH)
Digital Shareables on Suicide Prevention (NIMH)
Digital Shareables on Coping with COVID-19 (NIMH)
NIMH Director’s Messages about COVID-19 (NIMH)
NIMH Director’s Messages about Suicide (NIMH)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 16th in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
Using AI to Advance Understanding of Long COVID Syndrome
Posted on by Lawrence Tabak, D.D.S., Ph.D.

The COVID-19 pandemic continues to present considerable public health challenges in the United States and around the globe. One of the most puzzling is why many people who get over an initial and often relatively mild COVID illness later develop new and potentially debilitating symptoms. These symptoms run the gamut including fatigue, shortness of breath, brain fog, anxiety, and gastrointestinal trouble.
People understandably want answers to help them manage this complex condition referred to as Long COVID syndrome. But because Long COVID is so variable from person to person, it’s extremely difficult to work backwards and determine what these people had in common that might have made them susceptible to Long COVID. The variability also makes it difficult to identify all those who have Long COVID, whether they realize it or not. But a recent study, published in the journal Lancet Digital Health, shows that a well-trained computer and its artificial intelligence can help.
Researchers found that computers, after scanning thousands of electronic health records (EHRs) from people with Long COVID, could reliably make the call. The results, though still preliminary and in need of further validation, point the way to developing a fast, easy-to-use computer algorithm to help determine whether a person with a positive COVID test is likely to battle Long COVID.
In this groundbreaking study, NIH-supported researchers led by Emily Pfaff, University of North Carolina, Chapel Hill, and Melissa Haendel, the University of Colorado Anschutz Medical Campus, Aurora, relied on machine learning. In machine learning, a computer sifts through vast amounts of data to look for patterns. One reason machine learning is so powerful is that it doesn’t require humans to tell the computer which features it should look for. As such, machine learning can pick up on subtle patterns that people would otherwise miss.
In this case, Pfaff, Haendel, and team decided to “train” their computer on EHRs from people who had reported a COVID-19 infection. (The records are de-identified to protect patient privacy.) The researchers found just what they needed in the National COVID Cohort Collaborative (N3C), a national, publicly available data resource sponsored by NIH’s National Center for Advancing Translational Sciences. It is part of NIH’s Researching COVID to Enhance Recovery (RECOVER) initiative, which aims to improve understanding of Long COVID.
The researchers defined a group of more than 1.5 million adults in N3C who either had been diagnosed with COVID-19 or had a record of a positive COVID-19 test at least 90 days prior. Next, they examined common features, including any doctor visits, diagnoses, or medications, from the group’s roughly 100,000 adults.
They fed that EHR data into a computer, along with health information from almost 600 patients who’d been seen at a Long COVID clinic. They developed three machine learning models: one to identify potential long COVID patients across the whole dataset and two others that focused separately on people who had or hadn’t been hospitalized.
All three models proved effective for identifying people with potential Long-COVID. Each of the models had an 85 percent or better discrimination threshold, indicating they are highly accurate. That’s important because, once researchers can identify those with Long COVID in a large database of people such as N3C, they can begin to ask and answer many critical questions about any differences in an individual’s risk factors or treatment that might explain why some get Long COVID and others don’t.
This new study is also an excellent example of N3C’s goal to assemble data from EHRs that enable researchers around the world to get rapid answers and seek effective interventions for COVID-19, including its long-term health effects. It’s also made important progress toward the urgent goal of the RECOVER initiative to identify people with or at risk for Long COVID who may be eligible to participate in clinical trials of promising new treatment approaches.
Long COVID remains a puzzling public health challenge. Another recent NIH study published in the journal Annals of Internal Medicine set out to identify people with symptoms of Long COVID, most of whom had recovered from mild-to-moderate COVID-19 [2]. More than half had signs of Long COVID. But, despite extensive testing, the NIH researchers were unable to pinpoint any underlying cause of the Long COVID symptoms in most cases.
So if you’d like to help researchers solve this puzzle, RECOVER is now enrolling adults and kids—including those who have and have not had COVID—at more than 80 study sites around the country.
References:
[1] Identifying who has long COVID in the USA: a machine learning approach using N3C data. Pfaff ER, Girvin AT, Bennett TD, Bhatia A, Brooks IM, Deer RR, Dekermanjian JP, Jolley SE, Kahn MG, Kostka K, McMurry JA, Moffitt R, Walden A, Chute CG, Haendel MA; N3C Consortium. Lancet Digit Health. 2022 May 16:S2589-7500(22)00048-6.
[2] A longitudinal study of COVID-19 sequelae and immunity: baseline findings. Sneller MC, Liang CJ, Marques AR, Chung JY, Shanbhag SM, Fontana JR, Raza H, Okeke O, Dewar RL, Higgins BP, Tolstenko K, Kwan RW, Gittens KR, Seamon CA, McCormack G, Shaw JS, Okpali GM, Law M, Trihemasava K, Kennedy BD, Shi V, Justement JS, Buckner CM, Blazkova J, Moir S, Chun TW, Lane HC. Ann Intern Med. 2022 May 24:M21-4905.
Links:
COVID-19 Research (NIH)
National COVID Cohort Collaborative (N3C) (National Center for Advancing Translational Sciences/NIH)
Emily Pfaff (University of North Carolina, Chapel Hill)
Melissa Haendel (University of Colorado, Aurora)
NIH Support: National Center for Advancing Translational Sciences; National Institute of General Medical Sciences; National Institute of Allergy and Infectious Diseases
Next Page