severe COVID-19
Study Shows Benefits of COVID-19 Vaccines and Boosters
Posted on by Lawrence Tabak, D.D.S., Ph.D.

As colder temperatures settle in and people spend more time gathered indoors, cases of COVID-19 and other respiratory illnesses almost certainly will rise. That’s why, along with scheduling your annual flu shot, it’s now recommended that those age 5 and up should get an updated COVID-19 booster shot [1,2]. Not only will these new boosters guard against the original strain of the coronavirus that started the pandemic, they will heighten your immunity to the Omicron variant and several of the subvariants that continue to circulate in the U.S. with devastating effects.
At last count, about 14.8 million people in the U.S.—including me—have rolled up their sleeves to receive an updated booster shot [3]. It’s a good start, but it also means that most Americans aren’t fully up to date on their COVID-19 vaccines. If you or your loved ones are among them, a new study may provide some needed encouragement to make an appointment at a nearby pharmacy or clinic to get boosted [4].
A team of NIH-supported researchers found a remarkably low incidence of severe COVID-19 illness last fall, winter, and spring among more than 1.6 million veterans who’d been vaccinated and boosted. Severe illness was also quite low in individuals without immune-compromising conditions.
These latest findings, published in the journal JAMA, come from a research group led by Dan Kelly, University of California, San Francisco. He and his team conducted their study drawing on existing health data from the Veterans Health Administration (VA) within a time window of July 2021 and May 2022.
They identified 1.6 million people who’d had a primary-care visit within the last two years and were fully vaccinated for COVID-19, which included receiving a booster shot. Almost three-quarters of those identified were 65 and older. Nearly all were male, and more than 70 percent had another pre-existing health condition that put them at greater risk of becoming seriously ill from a COVID-19 infection.
Over a 24-week follow-up period for each fully vaccinated individual, 125 per 10,000 people had a breakthrough infection. That’s about 1 percent. Just 8.9 in 10,000 fully vaccinated people—less than 0.1 percent—died or were hospitalized from COVID-19 pneumonia. Drilling down deeper into the data:
• Individuals with an immune-compromising condition had a very low rate of hospitalization or death. In this group, 39.6 per 10,000 people had a serious breakthrough infection. That translates to 0.3 percent.
• For people with other preexisting health conditions, including diabetes and heart disease, hospitalization or death totaled 0.07 percent, or 6.7 per 10,000 people.
• For otherwise healthy adults aged 65 and older, the incidence of hospitalization or death was 1.9 per 10,000 people, or 0.02 percent.
• For boosted participants 65 or younger with no high-risk conditions, hospitalization or death came to less than 1 per 10,000 people. That comes to less than 0.01 percent.
It’s worth noting that these results reflect a period when the Delta and Omicron variants were circulating, and available boosters still were based solely on the original variant. Heading into this winter, the hope is that the updated “bivalent” boosters from Pfizer and Moderna will offer even broader protection as this terrible virus continues to evolve.
The Centers for Disease Control and Prevention continues to recommend that everyone stay up to date with their COVID-19 vaccines. That means all adults and kids 5 and older are encouraged to get boosted if it has been at least two months since their last COVID-19 vaccine dose. For older people and those with other health conditions, it’s even more important given their elevated risk for severe illness.
What if you’ve had a COVID-19 infection recently? Getting vaccinated or boosted a few months after you’ve had a COVID-19 infection will offer you even better protection in the future.
So, if you are among the millions of Americans who’ve been vaccinated for COVID-19 but are now due for a booster, don’t delay. Get yourself boosted to protect your own health and the health of your loved ones as the holidays approach.
References:
[1] CDC recommends the first updated COVID-19 booster. Centers for Disease Control and Prevention. September 1, 2022.
[2] CDC expands updated COVID-19 vaccines to include children ages 5 through 11. Centers for Disease Control and Prevention, October 12, 2022.
[3] COVID-19 vaccinations in the United States. Centers for Disease Control and Prevention.
[4] Incidence of severe COVID-19 illness following vaccination and booster with BNT162b2, mRNA-1273, and Ad26.COV2.S vaccines. Kelly JD, Leonard S, Hoggatt KJ, Boscardin WJ, Lum EN, Moss-Vazquez TA, Andino R, Wong JK, Byers A, Bravata DM, Tien PC, Keyhani S. JAMA. 2022 Oct 11;328(14):1427-1437.
Links:
COVID-19 Research (NIH)
Dan Kelly (University of California, San Francisco)
NIH Support: National Institute of Allergy and Infectious Diseases
How Severe COVID-19 Can Tragically Lead to Lung Failure and Death
Posted on by Dr. Francis Collins
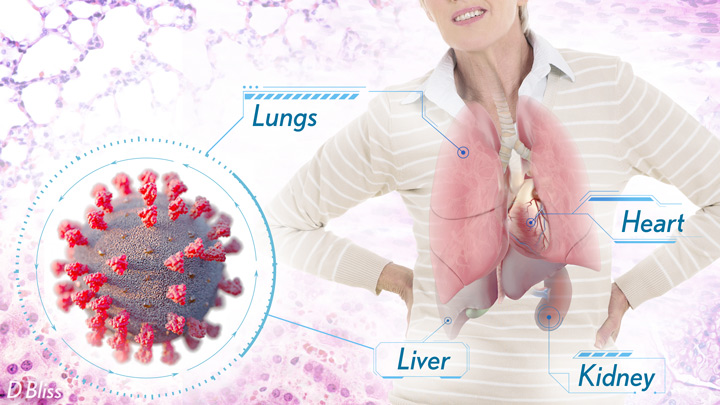
More than 3 million people around the world, now tragically including thousands every day in India, have lost their lives to severe COVID-19. Though incredible progress has been made in a little more than a year to develop effective vaccines, diagnostic tests, and treatments, there’s still much we don’t know about what precisely happens in the lungs and other parts of the body that leads to lethal outcomes.
Two recent studies in the journal Nature provide some of the most-detailed analyses yet about the effects on the human body of SARS-CoV-2, the coronavirus that causes COVID-19 [1,2]. The research shows that in people with advanced infections, SARS-CoV-2 often unleashes a devastating series of host events in the lungs prior to death. These events include runaway inflammation and rampant tissue destruction that the lungs cannot repair.
Both studies were supported by NIH. One comes from a team led by Benjamin Izar, Columbia University, New York. The other involves a group led by Aviv Regev, now at Genentech, and formerly at Broad Institute of MIT and Harvard, Cambridge, MA.
Each team analyzed samples of essential tissues gathered from COVID-19 patients shortly after their deaths. Izar’s team set up a rapid autopsy program to collect and freeze samples within hours of death. He and his team performed single-cell RNA sequencing on about 116,000 cells from the lung tissue of 19 men and women. Similarly, Regev’s team developed an autopsy biobank that included 420 total samples from 11 organ systems, which were used to generate multiple single-cell atlases of tissues from the lung, kidney, liver, and heart.
Izar’s team found that the lungs of people who died of COVID-19 were filled with immune cells called macrophages. While macrophages normally help to fight an infectious virus, they seemed in this case to produce a vicious cycle of severe inflammation that further damaged lung tissue. The researchers also discovered that the macrophages produced high levels of IL-1β, a type of small inflammatory protein called a cytokine. This suggests that drugs to reduce effects of IL-1β might have promise to control lung inflammation in the sickest patients.
As a person clears and recovers from a typical respiratory infection, such as the flu, the lung repairs the damage. But in severe COVID-19, both studies suggest this isn’t always possible. Not only does SARS-CoV-2 destroy cells within air sacs, called alveoli, that are essential for the exchange of oxygen and carbon dioxide, but the unchecked inflammation apparently also impairs remaining cells from repairing the damage. In fact, the lungs’ regenerative cells are suspended in a kind of reparative limbo, unable to complete the last steps needed to replace healthy alveolar tissue.
In both studies, the lung tissue also contained an unusually large number of fibroblast cells. Izar’s team went a step further to show increased numbers of a specific type of pathological fibroblast, which likely drives the rapid lung scarring (pulmonary fibrosis) seen in severe COVID-19. The findings point to specific fibroblast proteins that may serve as drug targets to block deleterious effects.
Regev’s team also describes how the virus affects other parts of the body. One surprising discovery was there was scant evidence of direct SARS-CoV-2 infection in the liver, kidney, or heart tissue of the deceased. Yet, a closer look heart tissue revealed widespread damage, documenting that many different coronary cell types had altered their genetic programs. It’s still to be determined if that’s because the virus had already been cleared from the heart prior to death. Alternatively, the heart damage might not be caused directly by SARS-CoV-2, and may arise from secondary immune and/or metabolic disruptions.
Together, these two studies provide clearer pictures of the pathology in the most severe and lethal cases of COVID-19. The data from these cell atlases has been made freely available for other researchers around the world to explore and analyze. The hope is that these vast data sets, together with future analyses and studies of people who’ve tragically lost their lives to this pandemic, will improve our understanding of long-term complications in patients who’ve survived. They also will now serve as an important foundational resource for the development of promising therapies, with the goal of preventing future complications and deaths due to COVID-19.
References:
[1] A molecular single-cell lung atlas of lethal COVID-19. Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, Katsyv I, Rendeiro AF, Amin AD, Schapiro D, Frangieh CJ, Luoma AM, Filliol A, Fang Y, Ravichandran H, Clausi MG, Alba GA, Rogava M, Chen SW, Ho P, Montoro DT, Kornberg AE, Han AS, Bakhoum MF, Anandasabapathy N, Suárez-Fariñas M, Bakhoum SF, Bram Y, Borczuk A, Guo XV, Lefkowitch JH, Marboe C, Lagana SM, Del Portillo A, Zorn E, Markowitz GS, Schwabe RF, Schwartz RE, Elemento O, Saqi A, Hibshoosh H, Que J, Izar B. Nature. 2021 Apr 29.
[2] COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Delorey TM, Ziegler CGK, Heimberg G, Normand R, Shalek AK, Villani AC, Rozenblatt-Rosen O, Regev A. et al. Nature. 2021 Apr 29.
Links:
COVID-19 Research (NIH)
Izar Lab (Columbia University, New York)
Aviv Regev (Genentech, South San Francisco, CA)
NIH Support: National Center for Advancing Translational Sciences; National Heart, Lung, and Blood Institute; National Cancer Institute; National Institute of Allergy and Infectious Diseases; National Institute of Diabetes and Digestive and Kidney Diseases; National Human Genome Research Institute; National Institute of Mental Health; National Institute on Alcohol Abuse and Alcoholism
Rogue Antibodies and Gene Mutations Explain Some Cases of Severe COVID-19
Posted on by Dr. Francis Collins
One of the many perplexing issues with COVID-19 is that it affects people so differently. That has researchers trying to explain why some folks bounce right back from the virus, or don’t even know they have it—while others become critically ill. Now, two NIH-funded studies suggest that one reason some otherwise healthy people become gravely ill may be previously unknown trouble spots in their immune systems, which hamper their ability to fight the virus.
According to the new findings in hundreds of racially diverse people with life-threatening COVID-19, a small percentage of people who suffer the most severe symptoms carry rare mutations in genes that disrupt their antiviral defenses. Another 10 percent with severe COVID-19 produce rogue “auto-antibodies,” which misguidedly disable a part of the immune system instead of attacking the virus.
Either way, the outcome is the same: the body has trouble fending off SARS-CoV-2, the novel coronavirus that causes COVID-19. The biological reason is there’s not enough of an assortment of signaling proteins, called type I interferons, that are crucial to detecting dangerous viruses like SARS-CoV-2 and sounding the alarm to prevent serious illness.
The research was led by Jean-Laurent Casanova, Howard Hughes Medical Institute and The Rockefeller University, New York; and the Imagine Institute, Necker Hospital, Paris. Casanova and his team began enrolling people with COVID-19 last February, with a particular interest in young adults battling severe illness. They were curious whether inherent weaknesses in their immune systems might explain their surprising vulnerability to the virus despite being otherwise young and healthy. Based on earlier findings in other infectious illnesses, they were especially interested in a set of 13 genes involved in interferon-driven immunity.
In their first study, published in the journal Science, researchers compared this set of genes in 659 patients with life-threatening COVID-19 to the same genes in 534 people with mild or asymptomatic COVID-19 [1]. It turned out that 23, or 3.5 percent, of people with severe COVID-19 indeed carried rare mutations in genes involved in producing antiviral interferons. Those unusual aberrations never turned up in people with milder disease. The researchers went on to show in lab studies that those genetic errors leave human cells more vulnerable to SARS-CoV-2 infection.
The discovery was certainly intriguing, but given the rarity of those mutations, it doesn’t explain most instances of severe COVID-19. Still, it did give Casanova’s team another idea. Perhaps some other people who suffer from severe COVID-19 lack interferons too, but for different reasons. Perhaps their bodies were producing rogue antibodies that were crippling their own antiviral defenses.
In their second study, also in Science, that’s exactly what researchers found in 101 of 987 (over 10 percent) patients from around the world with life-threatening COVID-19 [2]. In the bloodstreams of such individuals, they detected auto-antibodies against an assortment of interferon proteins. Those antibodies, which blocked the interferons’ antiviral activity, weren’t found in people with more mild cases of COVID-19.
Interestingly, the vast majority of patients with those harmful antibodies were men. The findings might help to explain the observation that men are at greater risk than women for developing severe COVID-19. The patients with auto-antibodies also were slightly older, with about half over the age of 65.
Many questions remain. For instance, it’s not yet clear what drives the production of those debilitating auto-antibodies. Might there be more mutations in antiviral defense-related genes that researchers have yet to discover? Is it possible that interferon treatment may help some people with severe COVID-19? Such treatment may be difficult in patients with auto-antibodies, although some clinical trials to explore this possibility already are underway.
The findings, if confirmed, have some potentially immediate implications. It’s possible that screening patients for the presence of damaging auto-antibodies might help to identify those at greater risk for progressing to severe disease. Treatments to remove those antibodies from the bloodstream or to boost antiviral defenses in other ways also may help. Ideally, it would be a good idea to make sure donated convalescent plasma now being tested in clinical trials as a treatment for severe COVID-19 doesn’t contain such disruptive auto-antibodies.
These new findings come from an international effort involving hundreds of scientists called the COVID Human Genetic Effort. Besides its ongoing efforts to understand severe COVID-19, Casanova says his team is also taking a look at the other side of the coin: how some people who’ve been exposed to severe COVID-19 in their own households manage to not get sick. A related international group called the COVID-19 Host Genetics Initiative is pursuing similar goals. Such insights will be invaluable as we continue to manage and treat COVID-19 patients in the future.
References:
[1] Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Gorochov G, Béziat V, Jouanguy E, Sancho-Shimizu V, Rice CM, Abel L, Notarangelo LD, Cobat A, Su HC, Casanova JL et al. Science. 2020 Sep 24:eabd4570. [Published online ahead of print.]
[2] Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Gorochov G, Jouanguy E, Rice CM, Cobat A, Notarangelo LD, Abel L, Su HC, Casanova JL et al. Science. 2020 Sep 24:eabd4585. [Published online ahead of print.]
Links:
Coronavirus (COVID-19) (NIH)
Interferons (Alpha, Beta) (NIH)
Interferons. Taylor MW. Viruses and Men: A History of Interactions. 2014 July 22. (Pubmed)
Video: Understanding the underlying genetics of COVID-19, Jean-Laurent Casanova (Youtube)
Jean-Laurent Casanova (The Rockefeller University, New York)
NIH Support: National Institute of Allergy and Infectious Diseases
Genes, Blood Type Tied to Risk of Severe COVID-19
Posted on by Dr. Francis Collins
Credit: National Institute of Allergy and Infectious Diseases, NIH
Many people who contract COVID-19 have only a mild illness, or sometimes no symptoms at all. But others develop respiratory failure that requires oxygen support or even a ventilator to help them recover [1]. It’s clear that this happens more often in men than in women, as well as in people who are older or who have chronic health conditions. But why does respiratory failure also sometimes occur in people who are young and seemingly healthy?
A new study suggests that part of the answer to this question may be found in the genes that each one of us carries [2]. While more research is needed to pinpoint the precise underlying genes and mechanisms responsible, a recent genome-wide association (GWAS) study, just published in the New England Journal of Medicine, finds that gene variants in two regions of the human genome are associated with severe COVID-19 and correspondingly carry a greater risk of COVID-19-related death.
The two stretches of DNA implicated as harboring risks for severe COVID-19 are known to carry some intriguing genes, including one that determines blood type and others that play various roles in the immune system. In fact, the findings suggest that people with blood type A face a 50 percent greater risk of needing oxygen support or a ventilator should they become infected with the novel coronavirus. In contrast, people with blood type O appear to have about a 50 percent reduced risk of severe COVID-19.
These new findings—the first to identify statistically significant susceptibility genes for the severity of COVID-19—come from a large research effort led by Andre Franke, a scientist at Christian-Albrecht-University, Kiel, Germany, along with Tom Karlsen, Oslo University Hospital Rikshospitalet, Norway. Their study included 1,980 people undergoing treatment for severe COVID-19 and respiratory failure at seven medical centers in Italy and Spain.
In search of gene variants that might play a role in the severe illness, the team analyzed patient genome data for more than 8.5 million so-called single-nucleotide polymorphisms, or SNPs. The vast majority of these single “letter” nucleotide substitutions found all across the genome are of no health significance, but they can help to pinpoint the locations of gene variants that turn up more often in association with particular traits or conditions—in this case, COVID-19-related respiratory failure. To find them, the researchers compared SNPs in people with severe COVID-19 to those in more than 1,200 healthy blood donors from the same population groups.
The analysis identified two places that turned up significantly more often in the individuals with severe COVID-19 than in the healthy folks. One of them is found on chromosome 3 and covers a cluster of six genes with potentially relevant functions. For instance, this portion of the genome encodes a transporter protein known to interact with angiotensin converting enzyme 2 (ACE2), the surface receptor that allows the novel coronavirus that causes COVID-19, SARS-CoV-2, to bind to and infect human cells. It also encodes a collection of chemokine receptors, which play a role in the immune response in the airways of our lungs.
The other association signal popped up on chromosome 9, right over the area of the genome that determines blood type. Whether you are classified as an A, B, AB, or O blood type, depends on how your genes instruct your blood cells to produce (or not produce) a certain set of proteins. The researchers did find evidence suggesting a relationship between blood type and COVID-19 risk. They noted that this area also includes a genetic variant associated with increased levels of interleukin-6, which plays a role in inflammation and may have implications for COVID-19 as well.
These findings, completed in two months under very difficult clinical conditions, clearly warrant further study to understand the implications more fully. Indeed, Franke, Karlsen, and many of their colleagues are part of the COVID-19 Host Genetics Initiative, an ongoing international collaborative effort to learn the genetic determinants of COVID-19 susceptibility, severity, and outcomes. Some NIH research groups are taking part in the initiative, and they recently launched a study to look for informative gene variants in 5,000 COVID-19 patients in the United States and Canada.
The hope is that these and other findings yet to come will point the way to a more thorough understanding of the biology of COVID-19. They also suggest that a genetic test and a person’s blood type might provide useful tools for identifying those who may be at greater risk of serious illness.
References:
[1] Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Wu Z, McGoogan JM, et. al. 2020 Feb 24. [published online ahead of print]
[2] Genomewide association study of severe Covid-19 with respiratory failure. Ellinghaus D, Degenhardt F, et. a. NEJM. June 17, 2020.
Links:
The COVID-19 Host Genetics Initiative
Andre Franke (Christian-Albrechts-University of Kiel, Germany)
Tom Karlsen (Oslo University Hospital Rikshospitalet, Norway)

