flu
Experimental mRNA Vaccine May Protect Against All 20 Influenza Virus Subtypes
Posted on by Lawrence Tabak, D.D.S., Ph.D.

Flu season is now upon us, and protecting yourself and loved ones is still as easy as heading to the nearest pharmacy for your annual flu shot. These vaccines are formulated each year to protect against up to four circulating strains of influenza virus, and they generally do a good job of this. What they can’t do is prevent future outbreaks of more novel flu viruses that occasionally spill over from other species into humans, thereby avoiding a future influenza pandemic.
On this latter and more-challenging front, there’s some encouraging news that was published recently in the journal Science [1]. An NIH-funded team has developed a unique “universal flu vaccine” that, with one seasonal shot, that has the potential to build immune protection against any of the 20 known subtypes of influenza virus and protect against future outbreaks.
While this experimental flu vaccine hasn’t yet been tested in people, the concept has shown great promise in advanced pre-clinical studies. Human clinical trials will hopefully start in the coming year. The researchers don’t expect that this universal flu vaccine will prevent influenza infection altogether. But, like COVID-19 vaccines, the new flu vaccine should help to reduce severe influenza illnesses and deaths when a person does get sick.
So, how does one develop a 20-in-1“multivalent” flu vaccine? It turns out that the key is the same messenger RNA (mRNA) technology that’s enabled two of the safe and effective vaccines against COVID-19, which have been so instrumental in fighting the pandemic. This includes the latest boosters from both Pfizer and Moderna, which now offer updated protection against currently circulating Omicron variants.
While this isn’t the first attempt to develop a universal flu vaccine, past attempts had primarily focused on a limited number of conserved antigens. An antigen is a protein or other substance that produces an immune response. Conserved antigens are those that tend to stay the same over time.
Because conserved antigens will look similar in many different influenza viruses, the hope was that vaccines targeting a small number of them would afford some broad influenza protection. But the focus on a strategy involving few antigens was driven largely by practical limitations. Using traditional methods to produce vaccines by growing flu viruses in eggs and isolating proteins, it simply isn’t feasible to include more than about four targets.
That’s where recent advances in mRNA technology come in. What makes mRNA so nifty for vaccines is that all you need to know is the letters, or sequence, that encodes the genetic material of a virus, including the sequences that get translated into proteins.
A research team led by Scott Hensley, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, recognized that the ease of designing and manufacturing mRNA vaccines opened the door to an alternate approach to developing a universal flu vaccine. Rather than limiting themselves to a few antigens, the researchers could make an all-in-one influenza vaccine, encoding antigens from every known influenza virus subtype.
Influenza vaccines generally target portions of a plentiful protein on the viral surface known as hemagglutinin (H). In earlier work, Hensley’s team, in collaboration with Perelman’s mRNA vaccine pioneer Drew Weissman, showed they could use mRNA technology to produce vaccines with H antigens from single influenza viruses [2, 3]. To protect the fragile mRNA molecules that encode a selected H antigen, researchers deliver them to cells inside well-tolerated microscopic lipid shells, or nanoparticles. The same is true of mRNA COVID-19 vaccines. In their earlier studies, the researchers found that when an mRNA vaccine aimed at one flu virus subtype was given to mice and ferrets in the lab, their cells made the encoded H antigen, eliciting protective antibodies.
In this latest study, they threw antigens from all 20 known flu viruses into the mix. This included H antigens from 18 known types of influenza A and two lineages of influenza B. The goal was to develop a vaccine that could teach the immune system to recognize and respond to any of them.
More study is needed, of course, but early indications are encouraging. The vaccine generated strong and broad antibody responses in animals. Importantly, it worked both in animals with no previous immunity to the flu and in those previously infected with flu viruses. That came as good news because past infections and resulting antibodies sometimes can interfere with the development of new antibodies against related viral subtypes.
In more good news, the researchers found that vaccinated mice and ferrets were protected against severe illness when later challenged with flu viruses. Those viruses included some that were closely matched to antigens in the vaccine, along with some that weren’t.
The findings offer proof-of-principle that mRNA vaccines containing a wide range of antigens can offer broad protection against influenza and likely other viruses as well, including the coronavirus strains responsible for COVID-19. The researchers report that they’re moving toward clinical trials in people, with the goal of beginning an early phase 1 trial in the coming year. The hope is that these developments—driven in part by technological advances and lessons learned over the course of the COVID-19 pandemic—will help to mitigate or perhaps even prevent future pandemics.
References:
[1] A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Arevalo CP, Bolton MJ, Le Sage V, Ye N, Furey C, Muramatsu H, Alameh MG, Pardi N, Drapeau EM, Parkhouse K, Garretson T, Morris JS, Moncla LH, Tam YK, Fan SHY, Lakdawala SS, Weissman D, Hensley SE. Science. 2022 Nov 25;378(6622):899-904.
[2] Nucleoside-modified mRNA vaccination partially overcomes maternal antibody inhibition of de novo immune responses in mice. Willis E, Pardi N, Parkhouse K, Mui BL, Tam YK, Weissman D, Hensley SE. Sci Transl Med. 2020 Jan 8;12(525):eaav5701.
[3] Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Pardi N, Parkhouse K, Kirkpatrick E, McMahon M, Zost SJ, Mui BL, Tam YK, Karikó K, Barbosa CJ, Madden TD, Hope MJ, Krammer F, Hensley SE, Weissman D. Nat Commun. 2018 Aug 22;9(1):3361.
Links:
Understanding Flu Viruses (Centers for Disease Control and Prevention, Atlanta)
COVID Research (NIH)
Decades in the Making: mRNA COVID-19 Vaccines (NIH)
Video: mRNA Flu Vaccines: Preventing the Next Pandemic (Penn Medicine, Philadelphia)
Scott Hensley (Perelman School of Medicine at the University of Pennsylvania, Philadelphia)
Weissman Lab (Perelman School of Medicine)
Video: The Story Behind mRNA COVID Vaccines: Katalin Karikó and Drew Weissman (Penn Medicine, Philadelphia)
NIH Support: National Institute for Allergy and Infectious Diseases
These Oddball Cells May Explain How Influenza Leads to Asthma
Posted on by Dr. Francis Collins
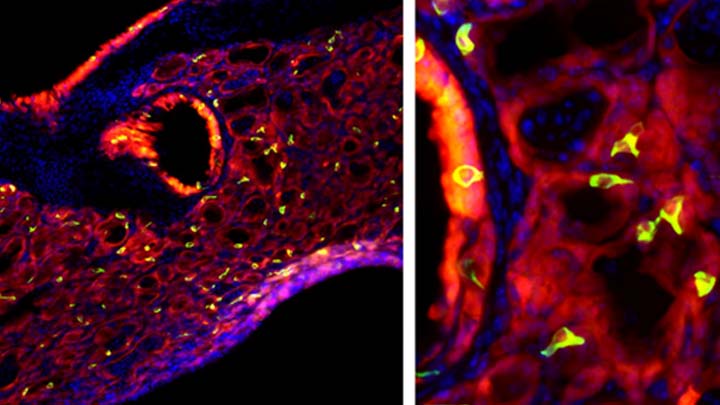
Most people who get the flu bounce right back in a week or two. But, for others, the respiratory infection is the beginning of lasting asthma-like symptoms. Though I had a flu shot, I had a pretty bad respiratory illness last fall, and since that time I’ve had exercise-induced asthma that has occasionally required an inhaler for treatment. What’s going on? An NIH-funded team now has evidence from mouse studies that such long-term consequences stem in part from a surprising source: previously unknown lung cells closely resembling those found in taste buds.
The image above shows the lungs of a mouse after a severe case of H1N1 influenza infection, a common type of seasonal flu. Notice the oddball cells (green) known as solitary chemosensory cells (SCCs). Those little-known cells display the very same chemical-sensing surface proteins found on the tongue, where they allow us to sense bitterness. What makes these images so interesting is, prior to infection, the healthy mouse lungs had no SCCs.
SCCs, sometimes called “tuft cells” or “brush cells” or “type II taste receptor cells”, were first described in the 1920s when a scientist noticed unusual looking cells in the intestinal lining [1] Over the years, such cells turned up in the epithelial linings of many parts of the body, including the pancreas, gallbladder, and nasal passages. Only much more recently did scientists realize that those cells were all essentially the same cell type. Owing to their sensory abilities, these epithelial cells act as a kind of lookout for signs of infection or injury.
This latest work on SCCs, published recently in the American Journal of Physiology–Lung Cellular and Molecular Physiology, adds to this understanding. It comes from a research team led by Andrew Vaughan, University of Pennsylvania School of Veterinary Medicine, Philadelphia [2].
As a post-doc, Vaughan and colleagues had discovered a new class of cells, called lineage-negative epithelial progenitors, that are involved in abnormal remodeling and regrowth of lung tissue after a serious respiratory infection [3]. Upon closer inspection, they noticed that the remodeling of lung tissue post-infection often was accompanied by sustained inflammation. What they didn’t know was why.
The team, including Noam Cohen of Penn’s Perelman School of Medicine and De’Broski Herbert, also of Penn Vet, noticed signs of an inflammatory immune response several weeks after an influenza infection. Such a response in other parts of the body is often associated with allergies and asthma. All were known to involve SCCs, and this begged the question: were SCCs also present in the lungs?
Further work showed not only were SCCs present in the lungs post-infection, they were interspersed across the tissue lining. When the researchers exposed the animals’ lungs to bitter compounds, the activated SCCs multiplied and triggered acute inflammation.
Vaughan’s team also found out something pretty cool. The SCCs arise from the very same lineage of epithelial progenitor cells that Vaughan had discovered as a post-doc. These progenitor cells produce cells involved in remodeling and repair of lung tissue after a serious lung infection.
Of course, mice aren’t people. The researchers now plan to look in human lung samples to confirm the presence of these cells following respiratory infections.
If confirmed, the new findings might help to explain why kids who acquire severe respiratory infections early in life are at greater risk of developing asthma. They suggest that treatments designed to control these SCCs might help to treat or perhaps even prevent lifelong respiratory problems. The hope is that ultimately it will help to keep more people breathing easier after a severe bout with the flu.
References:
[1] Closing in on a century-old mystery, scientists are figuring out what the body’s ‘tuft cells’ do. Leslie M. Science. 2019 Mar 28.
[2] Development of solitary chemosensory cells in the distal lung after severe influenza injury. Rane CK, Jackson SR, Pastore CF, Zhao G, Weiner AI, Patel NN, Herbert DR, Cohen NA, Vaughan AE. Am J Physiol Lung Cell Mol Physiol. 2019 Mar 25.
[3] Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, Matthay MA, Rock JR, Chapman HA. Nature. 2015 Jan 29;517(7536):621-625.
Links:
Asthma (National Heart, Lung, and Blood Institute/NIH)
Influenza (National Institute of Allergy and Infectious Diseases/NIH)
Vaughan Lab (University of Pennsylvania, Philadelphia)
Cohen Lab (University of Pennsylvania, Philadelphia)
Herbert Lab (University of Pennsylvania, Philadelphia)
NIH Support: National Heart, Lung, and Blood Institute; National Institute on Deafness and Other Communication Disorders
Looking to Llamas for New Ways to Fight the Flu
Posted on by Dr. Francis Collins
 Researchers are making tremendous strides toward developing better ways to reduce our risk of getting the flu. And one of the latest ideas for foiling the flu—a “gene mist” that could be sprayed into the nose—comes from a most surprising source: llamas.
Researchers are making tremendous strides toward developing better ways to reduce our risk of getting the flu. And one of the latest ideas for foiling the flu—a “gene mist” that could be sprayed into the nose—comes from a most surprising source: llamas.
Like humans and many other creatures, these fuzzy South American relatives of the camel produce immune molecules, called antibodies, in their blood when exposed to viruses and other foreign substances. Researchers speculated that because the llama’s antibodies are so much smaller than human antibodies, they might be easier to use therapeutically in fending off a wide range of flu viruses. This idea is now being leveraged to design a new type of gene therapy that may someday provide humans with broader protection against the flu [1].
Creative Minds: Does Human Immunity Change with the Seasons?
Posted on by Dr. Francis Collins
It’s an inescapable conclusion from the book of Ecclesiastes that’s become part of popular culture thanks to folk legends Pete Seeger and The Byrds: “To everything (turn, turn, turn), there is a season.” That’s certainly true of viral outbreaks, from the flu-causing influenza virus peaking each year in the winter to polio outbreaks often rising in the summer. What fascinates Micaela Martinez is, while those seasonal patterns of infection have been recognized for decades, nobody really knows why they occur.
Martinez, an infectious disease ecologist at Princeton University, Princeton, NJ, thinks colder weather conditions and the tendency for humans to stay together indoors in winter surely play a role. But she also thinks an important part of the answer might be found in a place most hadn’t thought to look: seasonal changes in the human immune system. Martinez recently received an NIH Director’s 2016 Early Independence Award to explore fluctuations in the body’s biological rhythms over the course of the year and their potential influence on our health.
Birth Year Predicts Bird Flu Risk
Posted on by Dr. Francis Collins

Caption: Birth years of people in China who contracted H7N9 avian flu from 1997-2015 (left); birth years of people in Cambodia, China, Egypt, Indonesia, Thailand, and Vietnam who contracted H5N1 avian flu from 1997-2015 (right).
Source: Adapted from Science. 2016 Nov 11;354(6313):722-726.
You probably can’t remember the first time you came down with the flu as a kid. But new evidence indicates that the human immune system never forgets its first encounters with an influenza virus, possibly even using that immunological “memory” to protect against future infections by novel strains of avian influenza, or bird flu.
In a study that looked at cases of bird flu in six countries in Asia and the Middle East between 1997 and 2015, an NIH-supported research team found that people born before 1968 were at lower risk of becoming seriously ill or dying from the H5N1 strain of the bird flu virus than were those born afterwards [1]. Just the opposite was true of another emerging strain of bird flu. People born before 1968 were at greater risk of becoming seriously ill or dying of H7N9, while those born after that date were more often protected.
Taking a Snapshot of the Human Immune System
Posted on by Dr. Francis Collins
Source: National Cancer Institute Visuals Online, NIH
There are numerous tests to gauge the health of your heart. But no such widely accepted test exists for the many parts of the immune system. How can we tell if the immune system is strong or weak? Or quantify how badly it’s malfunctioning when we suffer from asthma, allergies, or arthritis?
A team led by scientists at Stanford University has taken the first steps toward creating such a test—by taking “snapshots” of the immune system.
Before we talk about what they did, let me review how the immune system protects us against disease. The innate immune system is like a standing army that defends us against invading microbes. But the innate system has no memory. It doesn’t recognize the invaders more quickly if they return. This is the job of the adaptive immune system—B and T cells. These cells not only remember invaders; they’re able to adapt their weapons—antibodies and T-cell receptors—to make them more effective. Think of them as the Special Forces.
How Influenza Pandemics Occur
Posted on by Dr. Francis Collins
| Credit: National Institute of Allergy and Infectious Diseases, NIH |
Flu season is upon us! Check out this NIH video to see how these pandemics emerge and spread new flu viruses around the globe.



