yeast
How to Make Biopharmaceuticals Quickly in Small Batches
Posted on by Dr. Francis Collins

Caption: InSCyT system. Image shows (1) production module, (2) purification module, and (3) formulation module.
Credit: Felice Frankel Daniloff, Massachusetts Institute of Technology, Cambridge
Today, vaccines and other protein-based biologic drugs are typically made in large, dedicated manufacturing facilities. But that doesn’t always fit the need, and it could one day change. A team of researchers has engineered a miniaturized biopharmaceutical “factory” that could fit on a dining room table and produce hundreds to thousands of doses of a needed treatment in about three days.
As published recently in the journal Nature Biotechnology, this on-demand manufacturing system is called Integrated Scalable Cyto-Technology (InSCyT). It is fully automated and can be readily reconfigured to produce virtually any approved or experimental vaccine, hormone, replacement enzyme, antibody, or other biopharmaceutical. With further improvements and testing, InSCyT promises to give researchers and health care providers easy access to specialty biologics needed to treat rare diseases, as well as treatments for combating infectious disease outbreaks in remote towns or villages around the globe.
An Architectural Guide to the Nuclear Pore Complex
Posted on by Dr. Francis Collins
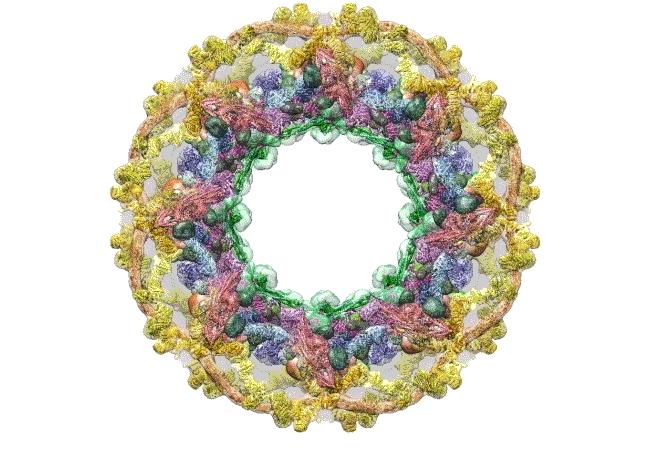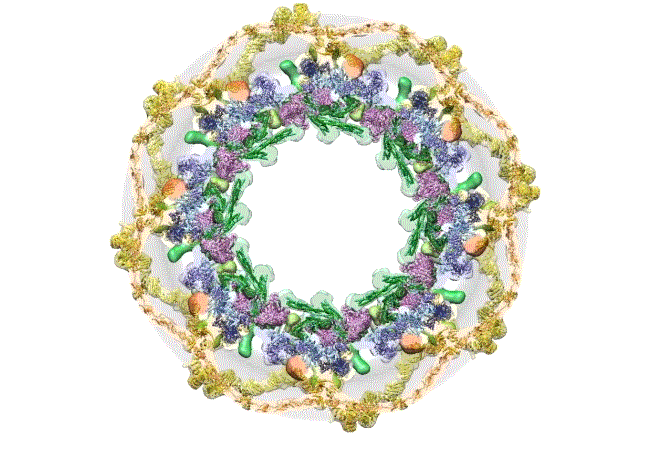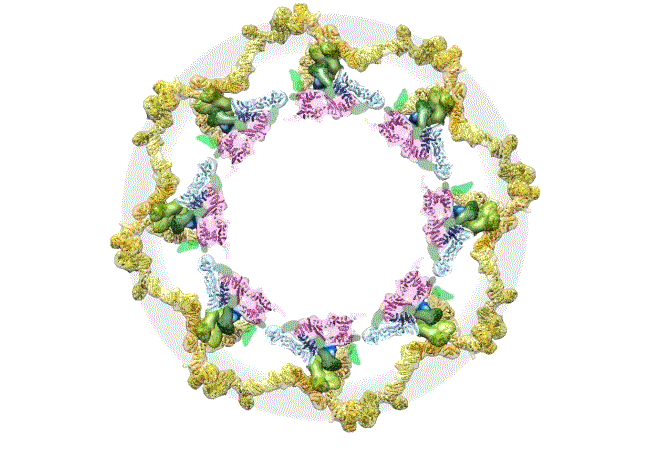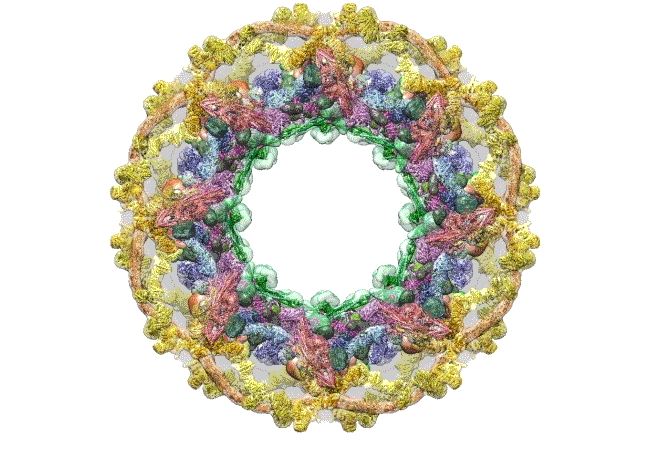 Credit: The Rockefeller University, New York
Credit: The Rockefeller University, New York
Sixty years ago, folk singer Pete Seeger recorded a song about helping those in need. The song starts like this: “Oh, had I a golden thread/And a needle so fine/I’d weave a magic strand/Of rainbow design.” In this brief animation, it seems like a golden thread and a needle are fast at work. But this rainbow design helps to answer a longstanding need for cell biologists: a comprehensive model of the thousands of pores embedded in the double-membrane barrier, or nuclear envelope, that divides the nucleus and its DNA from the rest of the cell.
These channels, called nuclear pore complexes (NPCs), are essential for life, tightly controlling which large macromolecules get in or out of the nucleus. Such activities include allowing vital proteins to enter the nucleus, blocking out harmful viruses, and shuttling messenger RNAs from the nucleus to the cytoplasm, where they are translated into proteins.
This computer simulation starts with an overhead view of the fully formed NPC structure. From this angle, the pore membrane (gray) appears to be at the base and is embroidered in four rings that are the channel’s main architectural support beams. There’s the cytoplasmic outer ring (yellow), the inner rings (purple, blue), the membrane ring (brown), and the nucleoplasmic outer ring (yellow). Each color represents different protein complexes, not rings per se, and the hole in the middle is the central channel through which molecules are transported. Filling the hole is a selective gating mechanism made of disordered protein (anchored to green) that helps to get the right molecules in and out.
Creative Minds: Using Machine Learning to Understand Genome Function
Posted on by Dr. Francis Collins
Science has always fascinated Anshul Kundaje, whether it was biology, physics, or chemistry. When he left his home country of India to pursue graduate studies in electrical engineering at Columbia University, New York, his plan was to focus on telecommunications and computer networks. But a course in computational genomics during his first semester showed him he could follow his interest in computing without giving up his love for biology.
Now an assistant professor of genetics and computer science at Stanford University, Palo Alto, CA, Kundaje has received a 2016 NIH Director’s New Innovator Award to explore not just how the human genome sequence encodes function, but also why it functions in the way that it does. Kundaje even envisions a time when it might be possible to use sophisticated computational approaches to predict the genomic basis of many human diseases.
Yeast Reveals New Drug Target for Parkinson’s
Posted on by Dr. Francis Collins

Credit: Daniel Tardiff, Whitehead Institute
Many progressive neurodegenerative disorders like Alzheimer’s, Huntington’s, and Parkinson’s disease, are characterized by abnormal clumps of proteins that clog up the cell and disrupt normal cellular functions. But it’s difficult to study these complex disease processes directly in the brain—so NIH-funded researchers, led by a team at the Whitehead Institute for Biomedical Research, Cambridge, MA, have turned to yeast for help.
Now, it may sound odd to study a brain disease in yeast, a microorganism long used in baking and brewing. After all, the brain is made up of billions of cells of many different types, while yeast grows as a single cell. But because the processes of protein production are generally conserved from yeast to humans, we can use this infinitely simpler organism to figure out what the proteins clumps are doing and test various drug candidates to halt the damage.



