technology
Clinical Center Doctors Testing 3D-Printed Miniature Ventilator
Posted on by James K. Gilman, MD, NIH Clinical Center

Here at the NIH Clinical Center, we are proud to be considered a world-renowned research hospital that provides hope through pioneering clinical research to improve human health. But what you may not know is that our doctors are constantly partnering with public and private sectors to come up with innovative technologies that will help to advance health outcomes.
I’m excited to bring to you a story that is perfect example of the ingenuity of our NIH doctors working with global strategic partners to create potentially life-saving technologies. This story begins during the COVID-19 pandemic with the global shortage of ventilators to help patients breathe. Hospitals had a profound need for inexpensive, easy-to-use, rapidly mass-produced resuscitation devices that could be quickly distributed in areas of critical need.
Through strategic partnerships, our Clinical Center doctors learned about and joined an international group of engineers, physicians, respiratory therapists, and patient advocates using their engineering skills to create a ventilator that was functional, affordable, and intuitive. After several iterations and bench testing, they devised a user-friendly ventilator.
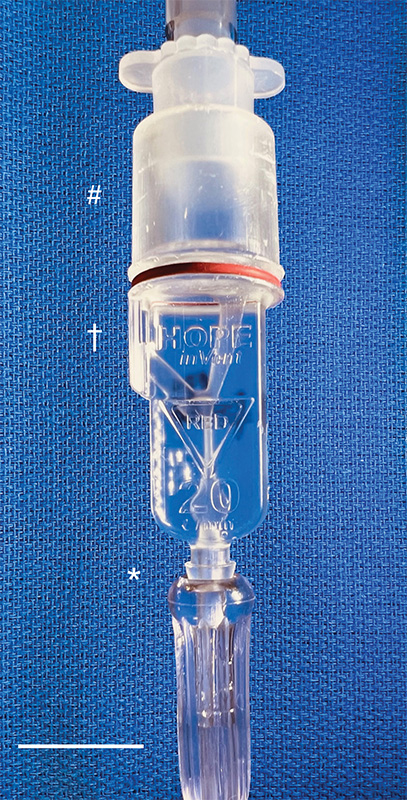
Then, with the assistance of 3D-printing technology, they improved the original design and did something pretty incredible: the team created the smallest single-patient ventilator seen to date. The device is just 2.4 centimeters (about 1 inch) in diameter with a length of 7.4 centimeters (about 3 inches).
A typical ventilator in a hospital obviously is much larger and has a bellows system. It fills with oxygen and then forces it into the lungs followed by the patient passively exhaling. These systems have multiple moving parts, valves, hoses, and electronic or mechanical controls to manage all aspects of the oxygen flow into the lungs.
But our miniature, 3D-printed ventilator is single use, disposable, and has no moving parts. It’s based on principles of fluidics to ventilate patients by automatically oscillating between forced inspiration and assisted expiration as airway pressure changes. It requires only a continuous supply of pressurized oxygen.
The possibilities of this 3D-printed miniature ventilator are broad. The ventilators could be easily used in emergency transport, potentially treating battlefield casualties or responding to disasters and mass casualty events like earthquakes.
While refining a concept is important, the key is converting it to actual use, which our doctors are doing admirably in their preclinical and clinical studies. NIH’s William Pritchard, Andrew Mannes, Brad Wood, John Karanian, Ivane Bakhutashvili, Matthew Starost, David Eckstein, and medical student Sheridan Reed studied and have already tested the ventilators in swine with acute lung injury, a common severe outcome in a number of respiratory threats including COVID-19.
In the study, the doctors tested three versions of the device built to correspond to mild, moderate, and severe lung injury. The respirators provided adequate support for moderate and mild lung injuries, and the doctors recall how amazing it was initially to witness a 190-pound swine ventilated by this miniature ventilator.
The doctors believe that the 3D-printed miniature ventilator is a potential “game changer” from start to finish since it is lifesaving, small, simple to use, can be easily and inexpensively printed and stored, and does not require additional maintenance. They recently published their preclinical trial results in the journal Science Translational Medicine [1].
The NIH team is preparing to initiate first-in-human trials here at the Clinical Center in the coming months. Perhaps, in the not-too-distant future, a device designed to help people breathe could fit into your pocket next to your phone and keys.
Reference:
[1] In-line miniature 3D-printed pressure-cycled ventilator maintains respiratory homeostasis in swine with induced acute pulmonary injury. Pritchard WF, Karanian JW, Jung C, Bakhutashvili I, Reed SL, Starost MF, Froelke BR, Barnes TR, Stevenson D, Mendoza A, Eckstein DJ, Wood BJ, Walsh BK, Mannes AJ. Sci Transl Med. 2022 Oct 12;14(666):eabm8351.
Links:
Clinical Center (NIH)
Andrew Mannes (Clinical Center)
Bradford Wood (Clinical Center)
David Eckstein (Clinical Center)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 21st in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
Swimming with the High-Tech Sharks to Improve COVID-19 Testing
Posted on by Dr. Francis Collins

So much has been reported over the past six months about testing for coronavirus disease 2019 (COVID-19) that keeping up with the issue can be a real challenge. To discuss the latest progress on new technologies for SARS-CoV-2 diagnostic testing in the United States, I spoke recently with NIH’s Dr. Bruce Tromberg, director of the National Institute of Biomedical Imaging and Bioengineering (NIBIB). Not only does Bruce run a busy NIH institute, he is helping to coordinate the national response for expanded testing during the COVID-19 pandemic.
Bruce also has a leading role in one of NIH’s most-exciting new initiatives. It’s called the Rapid Acceleration of Diagnostics (RADx) initiative, and it is on the fast track to bolster the country’s diagnostic testing capacity within months. Here’s a condensed transcript of our chat, which took place via videoconference, with Bruce linking in from Bethesda, MD and me from my home in Chevy Chase, MD:
Collins: Let’s start with how many COVID-19 tests are being done right now per day in the United States. By that, I’m referring to testing for the presence of the novel coronavirus, not antibodies as a sign of a previous infection.
Tromberg: The numbers fluctuate—anywhere from around 400,000 to 900,000 tests per day. So, the national capacity, with all these complex laboratory tests and emerging point-of-care assays, is getting close to 1 million a day. That’s substantially higher than in mid-April, when the nation was doing about 150,000 tests per day. But most testing is still being done in laboratories or complex facilities, and it can take a while for those tests to be run and for people to get answers. What we’d like to have are more convenient tests. We’d like to have tests that people can have at the point of care, where you get an answer on the spot and very quickly, or tests that can be performed easily in their homes.
Collins: Yes, we’d all love to have point-of-care tests for COVID-19. And there are some out there already. Every time I go to the White House, they have this gadget, called Abbott ID Now, that gives a result in about 15 minutes. That sounds pretty good. Do we just need to make more of those machines to solve the problem?
Tromberg: Abbott ID Now is one of the first point-of-care technologies. It’s not complicated, so a specialized laboratory isn’t required to run them. That’s what makes Abbott ID Now very appealing, but its performance could be better. There’s a bit of a risk when it’s used in individuals for which you really need to know, with absolute certainty, if they have the virus or not. Those performance issues have created opportunities to build platforms that are better, faster, and possible for people to do on their own.
Collins: Congress provided a big infusion of resources last April to assist in the development of new diagnostic technologies for COVID-19. A lot of that infusion came to NIH, and, Bruce, you were asked to step in and make something amazing happen on a timetable that’s pretty breathtaking. It’s called the RADx Initiative. Tell us a little about that.
Tromberg: RADx is short for Rapid Acceleration of Diagnostics. The goal of the initiative is to make it possible for everyone to have access to diagnostic testing for COVID-19 as easily and quickly as possible. As we pivot to doing surveillance in large populations, we will need greater testing capacity to help optimize the management of each individual. So, that’s really the aim of RADx, or RADx-tech, which is a special flavor of RADx.
Collins: Right, the goal of RADx-tech, which you are overseeing, is to identify some of these exciting new technologies and help scale them up quickly to the point where they can help people across the nation. Could you give us some examples?
Tromberg: Sure. One general class of technologies is called a lateral flow assay. These tests are small enough to fit in your hand and come in a convenient container. Basically, you can use a swab from your oral cavity and place it on one of the pads, and then you add a little bit of solution. The actual assay itself has a membrane inside of a little plastic container. The fluid flows across the membrane, and there’s chemistry that goes on inside the container to detect, for example, genetic material from the coronavirus. So, it can tell you if there is a presence of virus inside the swab. It’s very quick and straightforward. A line will “light up” if virus is present.
Another type of lateral flow assay, also small enough to hold in your hand, looks for proteins on the surface of the virus. You don’t have to break up the virus particle itself, but in this specific example, what captures the virus in this membrane is what’s called an aptamer. An aptamer is similar to an antibody, except it’s made from nucleic acid. It’s designed to bind very tightly with any molecule of interest. If you put a saliva sample into this assay, it moves up the membrane and some chemistry takes place. And then, you’ll see a line appear if there’s presence of a virus.
Collins: You just said saliva. I think a lot of people would much prefer, if they had to provide a sample, to use saliva instead of having a swab stuck in their nose, especially if it has to go all the way to the back of the nose. Does saliva work?
Tromberg: We hope so. Right now, RADx-tech has at least nine companies that are in what we call phase one, which is a significant step towards commercialization. Of those companies, more than half are looking at saliva or other kinds of sampling that’s not sticking swabs way up into the nasal cavity.
Another type of test is a lateral flow assay that fits directly into a mobile device like a tablet. It has a separate lateral flow apparatus, which looks like an elongated zip drive, and it slides right into the tablet itself. It’s something that’s not complicated. It would be easy to do at home. But rather than watching for the presence of a reaction, you look for a light inside the tablet to say the result is ready. And then, there is another color of light that comes directly from the lateral flow strip, that’s an indicator that the virus is present.
One last example is a nucleic acid test. This rectangular, hand-held device (see photo), reminiscent of a computer disc, looks inside the virus to amplify small traces of its nucleic acid to detectable levels. It is completely self-contained. To find that technology today, you generally must go to complex laboratories where the test is done on big machines, operated in multiple steps. Efforts are being made to reduce the size and the complexity of these devices so they can move out to point of care, without sacrificing the performance that we expect from a laboratory-based device.
Collins: That’s totally cool. Is the nucleic-acid test device that you just mentioned made for one-time use, and then you throw it away?
Tromberg: That’s their business model right now. I should probably mention something about cost. For example, you can imagine scaling up lateral flow assays very quickly to make tens of millions of tests. The components are inexpensive, and the tests may cost just a few dollars to make.
If you’re throwing away a nucleic acid test with its more-expensive components, obviously, the cost will be higher. Right now, if you go to a laboratory for a nucleic acid test, the cost may be on the order of $40 or so. With these one-time-use nucleic acid tests, the aim is to scale up the manufacturing to produce larger volumes that will bring the cost down. The estimates are maybe $60 per test.
Collins: That needs to come down more, obviously. In the months ahead, we’re talking about testing millions of people, maybe even fairly often to make sure that they haven’t been infected by SARS-CoV-2, the novel coronavirus that causes COVID-19. Is frequent testing the kind of thing that you’d like to be able to do by next fall?
Tromberg: Yes, and I think that that really speaks to the diversity of the types of tests that we need. I think there is a market, or the capacity, for some of the more expensive tests, if they’re extremely accurate and convenient. So, the nucleic acid test may cost more, but it will give you an answer very quickly and with very high sensitivity. It’s also very convenient. But the performance of that test may be very different from a standard lateral flow assay. Those tests will be far more accessible and very, very inexpensive, but they may have a higher false negative rate. We envision that every test that comes out of our innovation funnel will have documentation about its best-use case.
Collins: You mentioned your innovation funnel, sometimes called a “shark tank.” Say a little more about the RADx-tech shark tank. Who gets into it, and what happens when they get there?
Tromberg: At NIH, we’re into processes, and NIBIB created a very effective one 13 years ago with the Point of Care Technology Research Network (POCTRN). We’ve now leveraged this network to focus almost exclusively on COVID testing. POCTRN has five sites in the US. All have core resources, personnel, and expertise that are contributing to RADx-tech. Those include the ability to validate tests independently, the ability to do clinical studies in real-world samples and patients, and the ability to analyze manufacturing and scale-up needs while creating a roadmap for every project team to follow.
We have more than 200 people around the country working day and night on this process. If anyone has an idea about a COVID-19 test, you can and apply for funding on the POCTRN website. Your application will be reviewed by a panel of 30 experts within a day and, if approved, will move to the next stage, which is the shark tank.
In the shark tank [also called phase zero], a team of experts will spend about 150 to 200 person-hours with you evaluating the strengths and weaknesses of your test technically, clinically, and commercially. From this careful analysis, a detailed proposal will be presented to a steering committee, then sent to NIH. If we think it’s a great idea, the project will enter what we call phase one, with considerable financial support and the expectation that the company will hit its validation milestones within a month.
Collins: How far have things progressed, given that you just started RADx on April 29?
Tromberg: We have almost 60 projects that have entered or emerged from this shark-tank stage. I’m expecting that we’ll have around 15 projects in the phase one stage this month, and it’s very exciting to see them move there. If they can reach their validation milestones in that first month, they will be eligible to move to phase two. It involves a much larger chunk of money, so companies can move into manufacturing and scale up for distribution. We’re hoping to have between five and 10 companies emerge over time from this innovation funnel. But, by the end of the summer, we’d like to see at least two come out with products that will make a difference.
Collins: Wow, that’s just a few months away. How will you can get there so fast?
Tromberg: Sure. Some companies are further along than others. I can think of one that is quite far along with an established platform concept. This company has lots of expertise and has raised lots of money. We may be able to give them the surge that they need, plus the additional support with regulatory issues, commercialization, and manufacturing, in that short period of time to go to market.
Complementing that work is another of our initiatives called Advanced Technology Platforms (RADx-ATP). It’s designed to scale up existing technologies. For example, I mentioned a one-time-use nucleic acid test. It still needs validation, emergency use authorization, a little bit of manufacturing optimization. But we have other platforms out there that are much closer to commercialization, and RADx-ATP could be very impactful in getting some of those technologies out earlier.
Collins: You mentioned RADx-ATP, and we’ve been talking about RADx-tech, which is your shark tank approach. But there are a couple of other RADx components. Say something about those, please.
Tromberg: Our centerpiece component for doing demonstration projects is called RADx-UP. This is an effort across NIH to provide cutting-edge testing technologies in underserved populations. If I’m allowed to be the interviewer and turn the tables, I might bounce the question back to you. This is where your thinking directly influenced the whole RADx portfolio. So, maybe you can tell us more.
Collins: I can try. It’s very clear that COVID-19 has hit certain populations particularly hard, especially African American and Hispanic communities. And yet, those communities often have the least access to testing, which is sort of upside-down. We want to help identify people who are infected quickly, do the quarantining, and prevent the infection from spreading. That has simply not worked very well in a lot of underserved communities.
With resources from Congress, we made it a very high priority to set up demonstration projects of these advanced technologies in communities that would benefit significantly from them. We’re trying to bring together two really important NIH priorities: technology development and addressing health disparities. I’ve got to say, at this particular moment, when we’re all really focused on the fact that our nation is still riddled with health disparities, health inequities, and even racism, this is a moment where we should be doing everything we can to try to take our scientific capabilities and apply them to finding solutions.
So, we’re all pretty excited about RADx-UP. But there’s one other RADx, and I’ll throw this one back to you. It’s called RADx-rad. What the heck is that, Bruce?
Tromberg: Well, RADx-rad is the home for the technologies that are really far forward and futuristic. These are the technologies that won’t quite be ready for the time pressure of the innovation funnel. But they’re fantastic ideas. They’re projects that may be non-traditional in terms of the application of technology. They have been generated largely by other NIH institutes and centers. They’re important ideas and projects that just need to be supported with a longer time-window of return. We don’t want to lose out on the energy and the ideas and the creativity of those concepts.
Collins: Right now, the focus is on COVID-19 and the need for testing, especially within this calendar year. We hope, by the end of 2020 or the early part of 2021, to have vaccines for COVID-19 ready to go. But, moving forward, there will be other events that will probably make us wish that we had point-of-care diagnostics. So, in the process of doing what you’re doing with all of these components, hopefully we’re also preparing for future challenges.
Bruce, you’re an optimistic guy. At the same time, we’ve got to be realistic. Around September, when schools and colleges are contemplating whether it’s safe to open up, what would we hope that RADx could contribute to make that a better outcome?
Tromberg: That’s a tough question to answer, but I have a lot of confidence in our process. I’m confident that we’re engaging the innovation and entrepreneurial community in such a way that a lot of these ideas will move out and give us better performing tests and more of them. A rough number that I like to think about is the capacity to test roughly 2 percent of the population, around 6 million people per day. I think we’ll hit that target by the end of the year.
I’d like to see testing technologies move away from being based predominantly in laboratories. I’d like to see them more accessible to people as technologies that they can use in their homes. We’re now doing so many things from home. We’re working from home, we’re talking from home, we get our entertainment from home. Home-based testing is really the direction a lot of healthcare is going. We need to have these technologies. I think the level of sophistication and performance that we’re hoping for is possible, and the innovation and entrepreneurial community is working extremely hard to make it happen. No one has really asked us to do anything of this scale before, and I like to compare it to our Super Bowl.
Collins: Well, this is one exciting Super Bowl, that’s for sure! You’ve applied the venture capitalist strategy to RADx of trying to discover what’s out there, while not being afraid to invest in risky endeavors. You’re figuring out how to help promising technologies take their best shot and fail early, if they’re going to fail. And for technologies that are further along, you give them the needed resources to advance to commercialization.
We have great hopes and expectations that RADx will make a real difference. What we’re doing here is not just about cool science, it’s also about saving lives. I want to thank you for your incredible dedication, and your intellectual and engineering contributions to this initiative, which make it one of the most exciting things that NIH is doing right now.
Tromberg: Thank you, Francis.
Links:
Coronavirus (COVID-19) (NIH)
Rapid Acceleration of Diagnostics (RADx)
“Social engineering and bioengineering together can thwart the COVID-19 pandemic,” Director’s Corner, National Institute of Biomedical Imaging and Bioengineering/NIH)
Video: RADx Tech and POCTRN: Diagnosing Disease-Delivering Health (NIBIB/NIH)
Giving Thanks for Biomedical Research
Posted on by Dr. Francis Collins
This Thanksgiving, Americans have an abundance of reasons to be grateful—loving family and good food often come to mind. Here’s one more to add to the list: exciting progress in biomedical research. To check out some of that progress, I encourage you to watch this short video, produced by NIH’s National Institute of Biomedical Imaging and Engineering (NIBIB), that showcases a few cool gadgets and devices now under development.
Among the technological innovations is a wearable ultrasound patch for monitoring blood pressure [1]. The patch was developed by a research team led by Sheng Xu and Chonghe Wang, University of California San Diego, La Jolla. When this small patch is worn on the neck, it measures blood pressure in the central arteries and veins by emitting continuous ultrasound waves.
Other great technologies featured in the video include:
• Laser-Powered Glucose Meter. Peter So and Jeon Woong Kang, researchers at Massachusetts Institute of Technology (MIT), Cambridge, and their collaborators at MIT and University of Missouri, Columbia have developed a laser-powered device that measures glucose through the skin [2]. They report that this device potentially could provide accurate, continuous glucose monitoring for people with diabetes without the painful finger pricks.
• 15-Second Breast Scanner. Lihong Wang, a researcher at California Institute of Technology, Pasadena, and colleagues have combined laser light and sound waves to create a rapid, noninvasive, painless breast scan. It can be performed while a woman rests comfortably on a table without the radiation or compression of a standard mammogram [3].
• White Blood Cell Counter. Carlos Castro-Gonzalez, then a postdoc at Massachusetts Institute of Technology, Cambridge, and colleagues developed a portable, non-invasive home monitor to count white blood cells as they pass through capillaries inside a finger [4]. The test, which takes about 1 minute, can be carried out at home, and will help those undergoing chemotherapy to determine whether their white cell count has dropped too low for the next dose, avoiding risk for treatment-compromising infections.
• Neural-Enabled Prosthetic Hand (NEPH). Ranu Jung, a researcher at Florida International University, Miami, and colleagues have developed a prosthetic hand that restores a sense of touch, grip, and finger control for amputees [5]. NEPH is a fully implantable, wirelessly controlled system that directly stimulates nerves. More than two years ago, the FDA approved a first-in-human trial of the NEPH system.
If you want to check out more taxpayer-supported innovations, take a look at NIBIB’s two previous videos from 2013 and 2018 As always, let me offer thanks to you from the NIH family—and from all Americans who care about the future of their health—for your continued support. Happy Thanksgiving!
References:
[1] Monitoring of the central blood pressure waveform via a conformal ultrasonic device. Wang C, Li X, Hu H, Zhang, L, Huang Z, Lin M, Zhang Z, Yun Z, Huang B, Gong H, Bhaskaran S, Gu Y, Makihata M, Guo Y, Lei Y, Chen Y, Wang C, Li Y, Zhang T, Chen Z, Pisano AP, Zhang L, Zhou Q, Xu S. Nature Biomedical Engineering. September 2018, 687-695.
[2] Evaluation of accuracy dependence of Raman spectroscopic models on the ratio of calibration and validation points for non-invasive glucose sensing. Singh SP, Mukherjee S, Galindo LH, So PTC, Dasari RR, Khan UZ, Kannan R, Upendran A, Kang JW. Anal Bioanal Chem. 2018 Oct;410(25):6469-6475.
[3] Single-breath-hold photoacoustic computed tomography of the breast. Lin L, Hu P, Shi J, Appleton CM, Maslov K, Li L, Zhang R, Wang LV. Nat Commun. 2018 Jun 15;9(1):2352.
[4] Non-invasive detection of severe neutropenia in chemotherapy patients by optical imaging of nailfold microcirculation. Bourquard A, Pablo-Trinidad A, Butterworth I, Sánchez-Ferro Á, Cerrato C, Humala K, Fabra Urdiola M, Del Rio C, Valles B, Tucker-Schwartz JM, Lee ES, Vakoc BJ9, Padera TP, Ledesma-Carbayo MJ, Chen YB, Hochberg EP, Gray ML, Castro-González C. Sci Rep. 2018 Mar 28;8(1):5301.
[5] Enhancing Sensorimotor Integration Using a Neural Enabled Prosthetic Hand System
Links:
Sheng Xu Lab (University of California San Diego, La Jolla)
So Lab (Massachusetts Institute of Technology, Cambridge)
Lihong Wang (California Institute of Technology, Pasadena)
Video: Lihong Wang: Better Cancer Screenings
Carlos Castro-Gonzalez (Madrid-MIT M + Visión Consortium, Cambridge, MA)
Video: Carlos Castro-Gonzalez (YouTube)
Ranu Jung (Florida International University, Miami)
Video: New Prosthetic System Restores Sense of Touch (Florida International)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Neurological Diseases and Stroke; National Heart, Lung, and Blood Institute; National Cancer Institute; Common Fund
Whole-Genome Sequencing Plus AI Yields Same-Day Genetic Diagnoses
Posted on by Dr. Francis Collins

Back in April 2003, when the international Human Genome Project successfully completed the first reference sequence of the human DNA blueprint, we were thrilled to have achieved that feat in just 13 years. Sure, the U.S. contribution to that first human reference sequence cost an estimated $400 million, but we knew (or at least we hoped) that the costs would come down quickly, and the speed would accelerate. How far we’ve come since then! A new study shows that whole genome sequencing—combined with artificial intelligence (AI)—can now be used to diagnose genetic diseases in seriously ill babies in less than 24 hours.
Take a moment to absorb this. I would submit that there is no other technology in the history of planet Earth that has experienced this degree of progress in speed and affordability. And, at the same time, DNA sequence technology has achieved spectacularly high levels of accuracy. The time-honored adage that you can only get two out of three for “faster, better, and cheaper” has been broken—all three have been dramatically enhanced by the advances of the last 16 years.
Rapid diagnosis is critical for infants born with mysterious conditions because it enables them to receive potentially life-saving interventions as soon as possible after birth. In a study in Science Translational Medicine, NIH-funded researchers describe development of a highly automated, genome-sequencing pipeline that’s capable of routinely delivering a diagnosis to anxious parents and health-care professionals dramatically earlier than typically has been possible [1].
While the cost of rapid DNA sequencing continues to fall, challenges remain in utilizing this valuable tool to make quick diagnostic decisions. In most clinical settings, the wait for whole-genome sequencing results still runs more than two weeks. Attempts to obtain faster results also have been labor intensive, requiring dedicated teams of experts to sift through the data, one sample at a time.
In the new study, a research team led by Stephen Kingsmore, Rady Children’s Institute for Genomic Medicine, San Diego, CA, describes a streamlined approach that accelerates every step in the process, making it possible to obtain whole-genome test results in a median time of about 20 hours and with much less manual labor. They propose that the system could deliver answers for 30 patients per week using a single genome sequencing instrument.
Here’s how it works: Instead of manually preparing blood samples, his team used special microbeads to isolate DNA much more rapidly with very little labor. The approach reduced the time for sample preparation from 10 hours to less than three. Then, using a state-of-the-art DNA sequencer, they sequence those samples to obtain good quality whole genome data in just 15.5 hours.
The next potentially time-consuming challenge is making sense of all that data. To speed up the analysis, Kingsmore’s team took advantage of a machine-learning system called MOON. The automated platform sifts through all the data using artificial intelligence to search for potentially disease-causing variants.
The researchers paired MOON with a clinical language processing system, which allowed them to extract relevant information from the child’s electronic health records within seconds. Teaming that patient-specific information with data on more than 13,000 known genetic diseases in the scientific literature, the machine-learning system could pick out a likely disease-causing mutation out of 4.5 million potential variants in an impressive 5 minutes or less!
To put the system to the test, the researchers first evaluated its ability to reach a correct diagnosis in a sample of 101 children with 105 previously diagnosed genetic diseases. In nearly every case, the automated diagnosis matched the opinions reached previously via the more lengthy and laborious manual interpretation of experts.
Next, the researchers tested the automated system in assisting diagnosis of seven seriously ill infants in the intensive care unit, and three previously diagnosed infants. They showed that their automated system could reach a diagnosis in less than 20 hours. That’s compared to the fastest manual approach, which typically took about 48 hours. The automated system also required about 90 percent less manpower.
The system nailed a rapid diagnosis for 3 of 7 infants without returning any false-positive results. Those diagnoses were made with an average time savings of more than 22 hours. In each case, the early diagnosis immediately influenced the treatment those children received. That’s key given that, for young children suffering from serious and unexplained symptoms such as seizures, metabolic abnormalities, or immunodeficiencies, time is of the essence.
Of course, artificial intelligence may never replace doctors and other healthcare providers. Kingsmore notes that 106 years after the invention of the autopilot, two pilots are still required to fly a commercial aircraft. Likewise, health care decisions based on genome interpretation also will continue to require the expertise of skilled physicians.
Still, such a rapid automated system will prove incredibly useful. For instance, this system can provide immediate provisional diagnosis, allowing the experts to focus their attention on more difficult unsolved cases or other needs. It may also prove useful in re-evaluating the evidence in the many cases in which manual interpretation by experts fails to provide an answer.
The automated system may also be useful for periodically reanalyzing data in the many cases that remain unsolved. Keeping up with such reanalysis is a particular challenge considering that researchers continue to discover hundreds of disease-associated genes and thousands of variants each and every year. The hope is that in the years ahead, the combination of whole genome sequencing, artificial intelligence, and expert care will make all the difference in the lives of many more seriously ill babies and their families.
Reference:
[1] Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Clark MM, Hildreth A, Batalov S, Ding Y, Chowdhury S, Watkins K, Ellsworth K, Camp B, Kint CI, Yacoubian C, Farnaes L, Bainbridge MN, Beebe C, Braun JJA, Bray M, Carroll J, Cakici JA, Caylor SA, Clarke C, Creed MP, Friedman J, Frith A, Gain R, Gaughran M, George S, Gilmer S, Gleeson J, Gore J, Grunenwald H, Hovey RL, Janes ML, Lin K, McDonagh PD, McBride K, Mulrooney P, Nahas S, Oh D, Oriol A, Puckett L, Rady Z, Reese MG, Ryu J, Salz L, Sanford E, Stewart L, Sweeney N, Tokita M, Van Der Kraan L, White S, Wigby K, Williams B, Wong T, Wright MS, Yamada C, Schols P, Reynders J, Hall K, Dimmock D, Veeraraghavan N, Defay T, Kingsmore SF. Sci Transl Med. 2019 Apr 24;11(489).
Links:
DNA Sequencing Fact Sheet (National Human Genome Research Institute/NIH)
Genomics and Medicine (NHGRI/NIH)
Genetic and Rare Disease Information Center (National Center for Advancing Translational Sciences/NIH)
Stephen Kingsmore (Rady Children’s Institute for Genomic Medicine, San Diego, CA)
NIH Support: National Institute of Child Health and Human Development; National Human Genome Research Institute; National Center for Advancing Translational Sciences
Measuring Brain Chemistry
Posted on by Dr. Francis Collins

Credit: From the American Chemical Society’s “Personal Stories of Discovery”
Serotonin is one of the chemical messengers that nerve cells in the brain use to communicate. Modifying serotonin levels is one way that antidepressant and anti-anxiety medications are thought to work and help people feel better. But the precise nature of serotonin’s role in the brain is largely unknown.
That’s why Anne Andrews set out in the mid-1990s as a fellow at NIH’s National Institute of Mental Health to explore changes in serotonin levels in the brains of anxious mice. But she quickly realized it wasn’t possible. The tools available for measuring serotonin—and most other neurochemicals in the brain—couldn’t offer the needed precision to conduct her studies.
Instead of giving up, Andrews did something about it. In the late 1990s, she began formulating an idea for a neural probe to make direct and precise measurements of brain chemistry. Her progress was initially slow, partly because the probe she envisioned was technologically ahead of its time. Now at the University of California, Los Angeles (UCLA) more than 15 years later, she’s nearly there. Buoyed by recent scientific breakthroughs, the right team to get the job done, and the support of a 2017 NIH Director’s Transformative Research Award, Andrews expects to have the first fully functional devices ready within the next two years.
Seven More Awesome Technologies Made Possible by Your Tax Dollars
Posted on by Dr. Francis Collins
We live in a world energized by technological advances, from that new app on your smartphone to drones and self-driving cars. As you can see from this video, NIH-supported researchers are also major contributors, developing a wide range of amazing biomedical technologies that offer tremendous potential to improve our health.
Produced by the NIH’s National Institute of Biomedical Imaging and Bioengineering (NIBIB), this video starts by showcasing some cool fluorescent markers that are custom-designed to light up specific cells in the body. This technology is already helping surgeons see and remove tumor cells with greater precision in people with head and neck cancer [1]. Further down the road, it might also be used to light up nerves, which can be very difficult to see—and spare—during operations for cancer and other conditions.
Other great things to come include:
- A wearable tattoo that detects alcohol levels in perspiration and wirelessly transmits the information to a smartphone.
- Flexible coils that produce high quality images during magnetic resonance imaging (MRI) [2-3]. In the future, these individualized, screen-printed coils may improve the comfort and decrease the scan times of people undergoing MRI, especially infants and small children.
- A time-release capsule filled with a star-shaped polymer containing the anti-malarial drug ivermectin. The capsule slowly dissolves in the stomach over two weeks, with the goal of reducing the need for daily doses of ivermectin to prevent malaria infections in at-risk people [4].
- A new radiotracer to detect prostate cancer that has spread to other parts of the body. Early clinical trial results show the radiotracer, made up of carrier molecules bonded tightly to a radioactive atom, appears to be safe and effective [5].
- A new supercooling technique that promises to extend the time that organs donated for transplantation can remain viable outside the body [6-7]. For example, current technology can preserve donated livers outside the body for just 24 hours. In animal studies, this new technique quadruples that storage time to up to four days.
- A wearable skin patch with dissolvable microneedles capable of effectively delivering an influenza vaccine. This painless technology, which has produced promising early results in humans, may offer a simple, affordable alternative to needle-and-syringe immunization [8].
If you like what you see here, be sure to check out this previous NIH video that shows six more awesome biomedical technologies that your tax dollars are helping to create. So, let me extend a big thanks to you from those of us at NIH—and from all Americans who care about the future of their health—for your strong, continued support!
References:
[1] Image-guided surgery in cancer: A strategy to reduce incidence of positive surgical margins. Wiley Interdiscip Rev Syst Biol Med. 2018 Feb 23.
[2] Screen-printed flexible MRI receive coils. Corea JR, Flynn AM, Lechêne B, Scott G, Reed GD, Shin PJ, Lustig M, Arias AC. Nat Commun. 2016 Mar 10;7:10839.
[3] Printed Receive Coils with High Acoustic Transparency for Magnetic Resonance Guided Focused Ultrasound. Corea J, Ye P, Seo D, Butts-Pauly K, Arias AC, Lustig M. Sci Rep. 2018 Feb 21;8(1):3392.
[4] Oral, ultra-long-lasting drug delivery: Application toward malaria elimination goals. Bellinger AM, Jafari M1, Grant TM, Zhang S, Slater HC, Wenger EA, Mo S, Lee YL, Mazdiyasni H, Kogan L, Barman R, Cleveland C, Booth L, Bensel T, Minahan D, Hurowitz HM, Tai T, Daily J, Nikolic B, Wood L, Eckhoff PA, Langer R, Traverso G. Sci Transl Med. 2016 Nov 16;8(365):365ra157.
[5] Clinical Translation of a Dual Integrin avb3– and Gastrin-Releasing Peptide Receptor–Targeting PET Radiotracer, 68Ga-BBN-RGD. Zhang J, Niu G, Lang L, Li F, Fan X, Yan X, Yao S, Yan W, Huo L, Chen L, Li Z, Zhu Z, Chen X. J Nucl Med. 2017 Feb;58(2):228-234.
[6] Supercooling enables long-term transplantation survival following 4 days of liver preservation. Berendsen TA, Bruinsma BG, Puts CF, Saeidi N, Usta OB, Uygun BE, Izamis ML, Toner M, Yarmush ML, Uygun K. Nat Med. 2014 Jul;20(7):790-793.
[7] The promise of organ and tissue preservation to transform medicine. Giwa S, Lewis JK, Alvarez L, Langer R, Roth AE, et a. Nat Biotechnol. 2017 Jun 7;35(6):530-542.
[8] The safety, immunogenicity, and acceptability of inactivated influenza vaccine delivered by microneedle patch (TIV-MNP 2015): a randomised, partly blinded, placebo-controlled, phase 1 trial. Rouphael NG, Paine M, Mosley R, Henry S, McAllister DV, Kalluri H, Pewin W, Frew PM, Yu T, Thornburg NJ, Kabbani S, Lai L, Vassilieva EV, Skountzou I, Compans RW, Mulligan MJ, Prausnitz MR; TIV-MNP 2015 Study Group.
Links:
National Institute of Biomedical Imaging and Bioengineering (NIH)
Center for Wearable Sensors (University of California, San Diego)
Hyperpolarized MRI Technology Resource Center (University of California, San Francisco)
Center for Engineering in Medicine (Massachusetts General Hospital, Boston)
Center for Drug Design, Development and Delivery (Georgia Tech University, Atlanta)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of Allergy and Infectious Diseases
Creative Minds: Giving Bacteria Needles to Fight Intestinal Disease
Posted on by Dr. Francis Collins
For Salmonella and many other disease-causing bacteria that find their way into our bodies, infection begins with a poke. That’s because these bad bugs are equipped with a needle-like protein filament that punctures the outer membrane of human cells and then, like a syringe, injects dozens of toxic proteins that help them replicate.
Cammie Lesser at Massachusetts General Hospital and Harvard Medical School, Cambridge, and her colleagues are now on a mission to bioengineer strains of bacteria that don’t cause disease to make these same syringes, called type III secretion systems. The goal is to use such “good” bacteria to deliver therapeutic molecules, rather than toxins, to human cells. Their first target is the gastrointestinal tract, where they hope to knock out hard-to-beat bacterial infections or to relieve the chronic inflammation that comes with inflammatory bowel disease (IBD).
Next Page




