microbiology
Celebrating 2019 Biomedical Breakthroughs
Posted on by Dr. Francis Collins

Happy New Year! As we say goodbye to the Teens, let’s take a look back at 2019 and some of the groundbreaking scientific discoveries that closed out this remarkable decade.
Each December, the reporters and editors at the journal Science select their breakthrough of the year, and the choice for 2019 is nothing less than spectacular: An international network of radio astronomers published the first image of a black hole, the long-theorized cosmic singularity where gravity is so strong that even light cannot escape [1]. This one resides in a galaxy 53 million light-years from Earth! (A light-year equals about 6 trillion miles.)
Though the competition was certainly stiff in 2019, the biomedical sciences were well represented among Science’s “runner-up” breakthroughs. They include three breakthroughs that have received NIH support. Let’s take a look at them:
In a first, drug treats most cases of cystic fibrosis: Last October, two international research teams reported the results from phase 3 clinical trials of the triple drug therapy Trikafta to treat cystic fibrosis (CF). Their data showed Trikafta effectively compensates for the effects of a mutation carried by about 90 percent of people born with CF. Upon reviewing these impressive data, the Food and Drug Administration (FDA) approved Trikafta, developed by Vertex Pharmaceuticals.
The approval of Trikafta was a wonderful day for me personally, having co-led the team that isolated the CF gene 30 years ago. A few years later, I wrote a song called “Dare to Dream” imagining that wonderful day when “the story of CF is history.” Though we’ve still got more work to do, we’re getting a lot closer to making that dream come true. Indeed, with the approval of Trikafta, most people with CF have for the first time ever a real chance at managing this genetic disease as a chronic condition over the course of their lives. That’s a tremendous accomplishment considering that few with CF lived beyond their teens as recently as the 1980s.
Such progress has been made possible by decades of work involving a vast number of researchers, many funded by NIH, as well as by more than two decades of visionary and collaborative efforts between the Cystic Fibrosis Foundation and Aurora Biosciences (now, Vertex) that built upon that fundamental knowledge of the responsible gene and its protein product. Not only did this innovative approach serve to accelerate the development of therapies for CF, it established a model that may inform efforts to develop therapies for other rare genetic diseases.
Hope for Ebola patients, at last: It was just six years ago that news of a major Ebola outbreak in West Africa sounded a global health emergency of the highest order. Ebola virus disease was then recognized as an untreatable, rapidly fatal illness for the majority of those who contracted it. Though international control efforts ultimately contained the spread of the virus in West Africa within about two years, over 28,600 cases had been confirmed leading to more than 11,000 deaths—marking the largest known Ebola outbreak in human history. Most recently, another major outbreak continues to wreak havoc in northeastern Democratic Republic of Congo (DRC), where violent civil unrest is greatly challenging public health control efforts.
As troubling as this news remains, 2019 brought a needed breakthrough for the millions of people living in areas susceptible to Ebola outbreaks. A randomized clinical trial in the DRC evaluated four different drugs for treating acutely infected individuals, including an antibody against the virus called mAb114, and a cocktail of anti-Ebola antibodies referred to as REGN-EB3. The trial’s preliminary data showed that about 70 percent of the patients who received either mAb114 or the REGN-EB3 antibody cocktail survived, compared with about half of those given either of the other two medicines.
So compelling were these preliminary results that the trial, co-sponsored by NIH’s National Institute of Allergy and Infectious Diseases (NIAID) and the DRC’s National Institute for Biomedical Research, was halted last August. The results were also promptly made public to help save lives and stem the latest outbreak. All Ebola patients in the DRC treatment centers now are treated with one or the other of these two options. The trial results were recently published.
The NIH-developed mAb114 antibody and the REGN-EB3 cocktail are the first therapeutics to be shown in a scientifically rigorous study to be effective at treating Ebola. This work also demonstrates that ethically sound clinical research can be conducted under difficult conditions in the midst of a disease outbreak. In fact, the halted study was named Pamoja Tulinde Maisha (PALM), which means “together save lives” in Kiswahili.
To top off the life-saving progress in 2019, the FDA just approved the first vaccine for Ebola. Called Ervebo (earlier rVSV-ZEBOV), this single-dose injectable vaccine is a non-infectious version of an animal virus that has been genetically engineered to carry a segment of a gene from the Zaire species of the Ebola virus—the virus responsible for the current DRC outbreak and the West Africa outbreak. Because the vaccine does not contain the whole Zaire virus, it can’t cause Ebola. Results from a large study in Guinea conducted by the WHO indicated that the vaccine offered substantial protection against Ebola virus disease. Ervebo, produced by Merck, has already been given to over 259,000 individuals as part of the response to the DRC outbreak. The NIH has supported numerous clinical trials of the vaccine, including an ongoing study in West Africa.
Microbes combat malnourishment: Researchers discovered a few years ago that abnormal microbial communities, or microbiomes, in the intestine appear to contribute to childhood malnutrition. An NIH-supported research team followed up on this lead with a study of kids in Bangladesh, and it published last July its groundbreaking finding: that foods formulated to repair the “gut microbiome” helped malnourished kids rebuild their health. The researchers were able to identify a network of 15 bacterial species that consistently interact in the gut microbiomes of Bangladeshi children. In this month-long study, this bacterial network helped the researchers characterize a child’s microbiome and/or its relative state of repair.
But a month isn’t long enough to determine how the new foods would help children grow and recover. The researchers are conducting a similar study that is much longer and larger. Globally, malnutrition affects an estimated 238 million children under the age 5, stunting their normal growth, compromising their health, and limiting their mental development. The hope is that these new foods and others adapted for use around the world soon will help many more kids grow up to be healthy adults.
Measles Resurgent: The staff at Science also listed their less-encouraging 2019 Breakdowns of the Year, and unfortunately the biomedical sciences made the cut with the return of measles in the U.S. Prior to 1963, when the measles vaccine was developed, 3 to 4 million Americans were sickened by measles each year. Each year about 500 children would die from measles, and many more would suffer lifelong complications. As more people were vaccinated, the incidence of measles plummeted. By the year 2000, the disease was even declared eliminated from the U.S.
But, as more parents have chosen not to vaccinate their children, driven by the now debunked claim that vaccines are connected to autism, measles has made a very preventable comeback. Last October, the Centers for Disease Control and Prevention (CDC) reported an estimated 1,250 measles cases in the United States at that point in 2019, surpassing the total number of cases reported annually in each of the past 25 years.
The good news is those numbers can be reduced if more people get the vaccine, which has been shown repeatedly in many large and rigorous studies to be safe and effective. The CDC recommends that children should receive their first dose by 12 to 15 months of age and a second dose between the ages of 4 and 6. Older people who’ve been vaccinated or have had the measles previously should consider being re-vaccinated, especially if they live in places with low vaccination rates or will be traveling to countries where measles are endemic.
Despite this public health breakdown, 2019 closed out a memorable decade of scientific discovery. The Twenties will build on discoveries made during the Teens and bring us even closer to an era of precision medicine to improve the lives of millions of Americans. So, onward to 2020—and happy New Year!
Reference:
[1] 2019 Breakthrough of the Year. Science, December 19, 2019.
NIH Support: These breakthroughs represent the culmination of years of research involving many investigators and the support of multiple NIH institutes.
How Mucus Tames Microbes
Posted on by Dr. Francis Collins

Most of us think of mucus as little more than slimy and somewhat yucky stuff that’s easily ignored until you come down with a cold like the one I just had. But, when it comes to our health, there’s much more to mucus than you might think.
Mucus covers the moist surfaces of the human body, including the eyes, nostrils, lungs, and gastrointestinal tract. In fact, the average person makes more than a liter of mucus each day! It houses trillions of microbes and serves as a first line of defense against the subset of those microorganisms that cause infections. For these reasons, NIH-funded researchers, led by Katharina Ribbeck, Massachusetts Institute of Technology, Cambridge, are out to gain a greater understanding of the biology of healthy mucus—and then possibly use that knowledge to develop new therapeutics.
Ribbeck’s team used a scanning electron microscope to take the image of mucus you see above. You’ll notice right away that mucus doesn’t look like simple slime at all. In fact, if you could zoom into this complex web, you’d discover it’s made up of mucin proteins and glycans, which are sugar molecules that resemble bottle brushes.
Ribbeck and her colleagues recently discovered that the glycans in healthy mucus play a long-overlooked role in “taming” bacteria that might make us ill [1]. This work builds on their previous findings that mucus interferes with bacterial behavior, preventing these bugs from attaching to surfaces and communicating with each other [2].
In their new study, published in Nature Microbiology, Ribbeck, lead author Kelsey Wheeler, and their colleagues studied mucus and its interactions with Pseudomonas aeruginosa. This bacterium is a common cause of serious lung infections in people with cystic fibrosis or compromised immune systems.
The researchers found that in the presence of glycans, P. aeruginosa was rendered less harmful and infectious. The bacteria also produced fewer toxins. The findings show that it isn’t just that microbes get trapped in a tangled web within mucus, but rather that glycans have a special ability to moderate the bugs’ behavior. The researchers also have evidence of similar interactions between mucus and other microorganisms, such as those responsible for yeast infections.
The new study highlights an intriguing strategy to tame, rather than kill, bacteria to manage infections. In fact, Ribbeck views mucus and its glycans as a therapeutic gold mine. She hopes to apply what she’s learned to develop artificial mucus as an anti-microbial therapeutic for use inside and outside the body. Not bad for a substance that you might have thought was nothing more than slimy stuff.
References:
[1] Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Wheeler KM, Cárcamo-Oyarce G, Turner BS, Dellos-Nolan S, Co JY, Lehoux S, Cummings RD, Wozniak DJ, Ribbeck K. Nat Microbiol. 2019 Oct 14.
[2] Mucins trigger dispersal of Pseudomonas aeruginosa biofilms. Co JY, Cárcamo-Oyarce, Billings N, Wheeler KM, Grindy SC, Holten-Andersen N, Ribbeck K. NPJ Biofilms Microbiomes. 2018 Oct 10;4:23.
Links:
Cystic Fibrosis (National Heart, Lung, and Blood Institute/NIH)
Video: Chemistry in Action—Katharina Ribbeck (YouTube)
Katharina Ribbeck (Massachusetts Institute of Technology, Cambridge)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Environmental Health Sciences; National Institute of General Medical Sciences; National Institute of Allergy and Infectious Diseases
Gut-Dwelling Bacterium Consumes Parkinson’s Drug
Posted on by Dr. Francis Collins

Scientists continue to uncover the many fascinating ways in which the trillions of microbes that inhabit the human body influence our health. Now comes yet another surprising discovery: a medicine-eating bacterium residing in the human gut that may affect how well someone responds to the most commonly prescribed drug for Parkinson’s disease.
There have been previous hints that gut microbes might influence the effectiveness of levodopa (L-dopa), which helps to ease the stiffness, rigidity, and slowness of movement associated with Parkinson’s disease. Now, in findings published in Science, an NIH-funded team has identified a specific, gut-dwelling bacterium that consumes L-dopa [1]. The scientists have also identified the bacterial genes and enzymes involved in the process.
Parkinson’s disease is a progressive neurodegenerative condition in which the dopamine-producing cells in a portion of the brain called the substantia nigra begin to sicken and die. Because these cells and their dopamine are critical for controlling movement, their death leads to the familiar tremor, difficulty moving, and the characteristic slow gait. As the disease progresses, cognitive and behavioral problems can take hold, including depression, personality shifts, and sleep disturbances.
For the 10 million people in the world now living with this neurodegenerative disorder, and for those who’ve gone before them, L-dopa has been for the last 50 years the mainstay of treatment to help alleviate those motor symptoms. The drug is a precursor of dopamine, and, unlike dopamine, it has the advantage of crossing the blood-brain barrier. Once inside the brain, an enzyme called DOPA decarboxylase converts L-dopa to dopamine.
Unfortunately, only a small fraction of L-dopa ever reaches the brain, contributing to big differences in the drug’s efficacy from person to person. Since the 1970s, researchers have suspected that these differences could be traced, in part, to microbes in the gut breaking down L-dopa before it gets to the brain.
To take a closer look in the new study, Vayu Maini Rekdal and Emily Balskus, Harvard University, Cambridge, MA, turned to data from the NIH-supported Human Microbiome Project (HMP). The project used DNA sequencing to identify and characterize the diverse collection of microbes that populate the healthy human body.
The researchers sifted through the HMP database for bacterial DNA sequences that appeared to encode an enzyme capable of converting L-dopa to dopamine. They found what they were looking for in a bacterial group known as Enterococcus, which often inhabits the human gastrointestinal tract.
Next, they tested the ability of seven representative Enterococcus strains to transform L-dopa. Only one fit the bill: a bacterium called Enterococcus faecalis, which commonly resides in a healthy gut microbiome. In their tests, this bacterium avidly consumed all the L-dopa, using its own version of a decarboxylase enzyme. When a specific gene in its genome was inactivated, E. faecalis stopped breaking down L-dopa.
These studies also revealed variability among human microbiome samples. In seven stool samples, the microbes tested didn’t consume L-dopa at all. But in 12 other samples, microbes consumed 25 to 98 percent of the L-dopa!
The researchers went on to find a strong association between the degree of L-dopa consumption and the abundance of E. faecalis in a particular microbiome sample. They also showed that adding E. faecalis to a sample that couldn’t consume L-dopa transformed it into one that could.
So how can this information be used to help people with Parkinson’s disease? Answers are already appearing. The researchers have found a small molecule that prevents the E. faecalis decarboxylase from modifying L-dopa—without harming the microbe and possibly destabilizing an otherwise healthy gut microbiome.
The finding suggests that the human gut microbiome might hold a key to predicting how well people with Parkinson’s disease will respond to L-dopa, and ultimately improving treatment outcomes. The finding also serves to remind us just how much the microbiome still has to tell us about human health and well-being.
Reference:
[1] Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Science. 2019 Jun 14;364(6445).
Links:
Parkinson’s Disease Information Page (National Institute of Neurological Disorders and Stroke/NIH)
Balskus Lab (Harvard University, Cambridge, MA)
NIH Support: National Institute of General Medical Sciences; National Heart, Lung, and Blood Institute
Some ‘Hospital-Acquired’ Infections Traced to Patient’s Own Microbiome
Posted on by Dr. Francis Collins
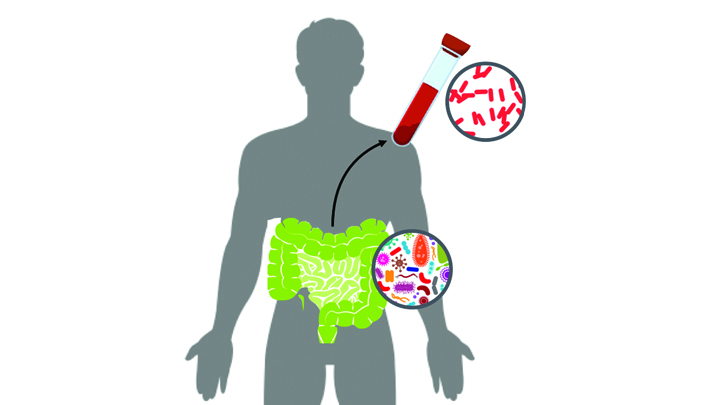
Caption: New computational tool determines whether a gut microbe is the source of a hospital-acquired bloodstream infection
Credit: Fiona Tamburini, Stanford University, Palo Alto, CA
While being cared for in the hospital, a disturbingly large number of people develop potentially life-threatening bloodstream infections. It’s been thought that most of the blame lies with microbes lurking on medical equipment, health-care professionals, or other patients and visitors. And certainly that is often true. But now an NIH-funded team has discovered that a significant fraction of these “hospital-acquired” infections may actually stem from a quite different source: the patient’s own body.
In a study of 30 bone-marrow transplant patients suffering from bloodstream infections, researchers used a newly developed computational tool called StrainSifter to match microbial DNA from close to one-third of the infections to bugs already living in the patients’ large intestines [1]. In contrast, the researchers found little DNA evidence to support the notion that such microbes were being passed around among patients.
A Bacterium Reaches Out and Grabs Some New DNA
Posted on by Dr. Francis Collins
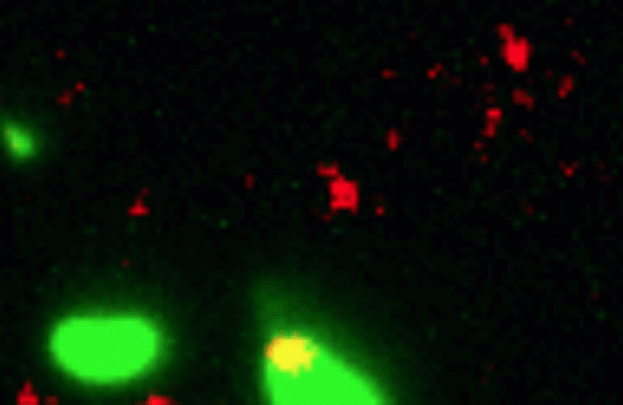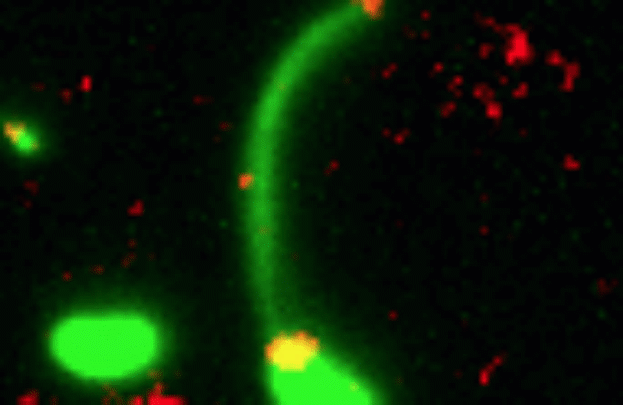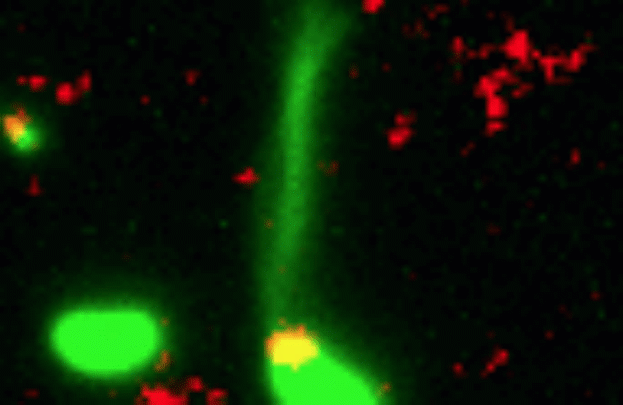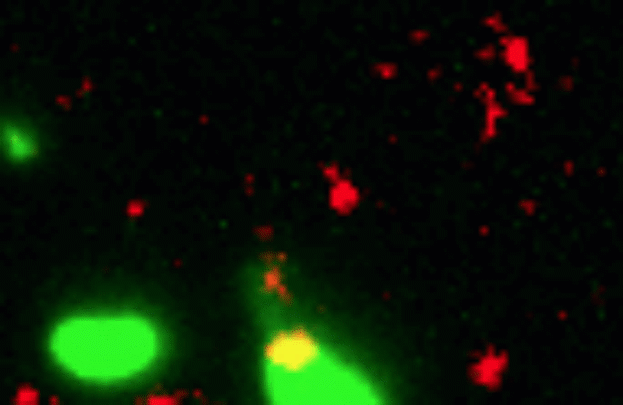
Credit: Dalia Lab, Indiana University, Bloomington
If you like comic book heroes, you’ll love this action-packed video of a microbe with a superpower reminiscent of a miniature Spiderman. Here, for the first time ever, scientists have captured in real-time—and in very cool detail—the important mechanism of horizontal gene transfer in bacteria.
Specifically, you see Vibrio cholerae, the water-dwelling bacterium that causes cholera, stretching out a hair-like appendage called a pilus (green) to snag a free snippet of DNA (red). After grabbing the DNA, V. cholerae swiftly retracts the pilus, threading the DNA fragment through a pore on the cell surface for stitching into its genome.
Creative Minds: Giving Bacteria Needles to Fight Intestinal Disease
Posted on by Dr. Francis Collins
For Salmonella and many other disease-causing bacteria that find their way into our bodies, infection begins with a poke. That’s because these bad bugs are equipped with a needle-like protein filament that punctures the outer membrane of human cells and then, like a syringe, injects dozens of toxic proteins that help them replicate.
Cammie Lesser at Massachusetts General Hospital and Harvard Medical School, Cambridge, and her colleagues are now on a mission to bioengineer strains of bacteria that don’t cause disease to make these same syringes, called type III secretion systems. The goal is to use such “good” bacteria to deliver therapeutic molecules, rather than toxins, to human cells. Their first target is the gastrointestinal tract, where they hope to knock out hard-to-beat bacterial infections or to relieve the chronic inflammation that comes with inflammatory bowel disease (IBD).
Snapshots of Life: Fighting Urinary Tract Infections
Posted on by Dr. Francis Collins

Source: Valerie O’Brien, Matthew Joens, Scott J. Hultgren, James A.J. Fitzpatrick, Washington University, St. Louis
For patients who’ve succeeded in knocking out a bad urinary tract infection (UTI) with antibiotic treatment, it’s frustrating to have that uncomfortable burning sensation flare back up. Researchers are hopeful that this striking work of science and art can help them better understand why severe UTIs leave people at greater risk of subsequent infection, as well as find ways to stop the vicious cycle.
Here you see the bladder (blue) of a laboratory mouse that was re-infected 24 hours earlier with the bacterium Escherichia coli (pink), a common cause of UTIs. White blood cells (yellow) reach out with what appear to be stringy extracellular traps to immobilize and kill the bacteria.
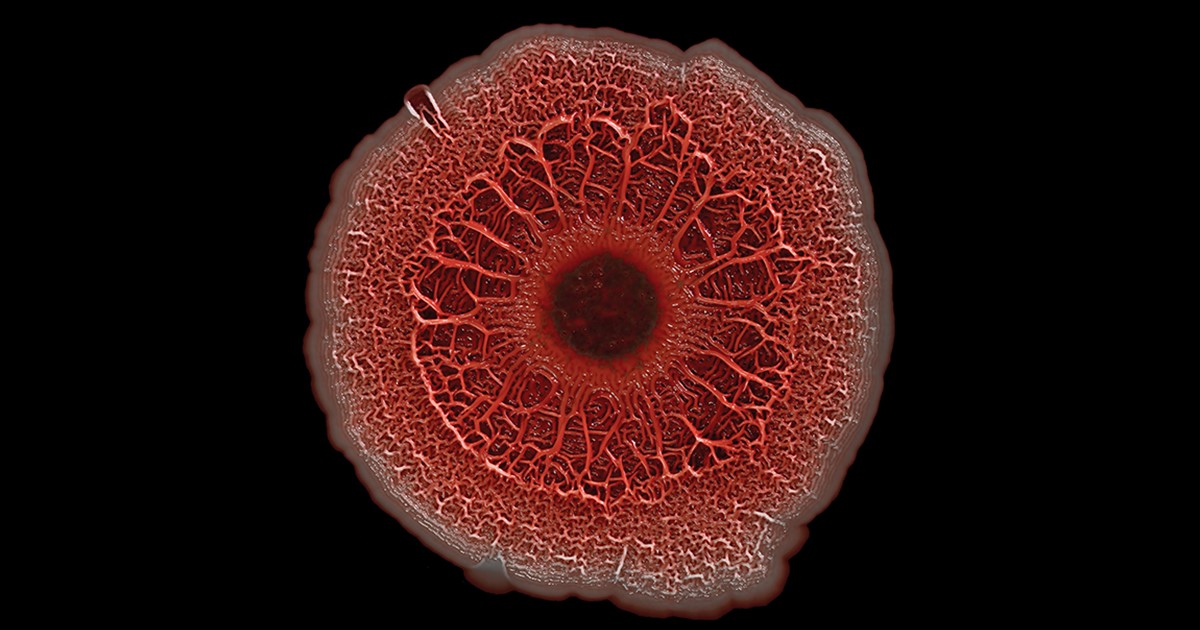
Snapshots of Life: Portrait of a Bacterial Biofilm
Posted on by Dr. Francis Collins
In nature, there is strength in numbers. Sometimes, those numbers also have their own unique beauty. That’s the story behind this image showing an intricate colony of millions of the single-celled bacterium Pseudomonas aeruginosa, a common culprit in the more than 700,000 hospital-acquired infections estimated to occur annually in the United States. [1]. The bacteria have self-organized into a sticky, mat-like colony called a biofilm, which allows them to cooperate with each other, adapt to changes in their environment, and ensure their survival.
In this image, the Pseudomonas biofilm has grown in a laboratory dish to about the size of a dime. Together, the millions of independent bacterial cells have created a tough extracellular matrix of secreted proteins, polysaccharide sugars, and even DNA that holds the biofilm together, stained in red. The darkened areas at the center come from the bacteria’s natural pigments.
Next Page


 Many people still regard bacteria and other microbes just as disease-causing germs. But it’s a lot more complicated than that. In fact, it’s become increasingly clear that the healthy human body is teeming with microorganisms, many of which play essential roles in our metabolism, our immune response, and even our mental health. We are not just an organism, we are a “superorganism” made up of human cells and microbial cells—and the microbes outnumber us! Fueling this new understanding is NIH’s Human Microbiome Project (HMP), a quest begun a decade ago to explore the microbial makeup of healthy Americans.
Many people still regard bacteria and other microbes just as disease-causing germs. But it’s a lot more complicated than that. In fact, it’s become increasingly clear that the healthy human body is teeming with microorganisms, many of which play essential roles in our metabolism, our immune response, and even our mental health. We are not just an organism, we are a “superorganism” made up of human cells and microbial cells—and the microbes outnumber us! Fueling this new understanding is NIH’s Human Microbiome Project (HMP), a quest begun a decade ago to explore the microbial makeup of healthy Americans.