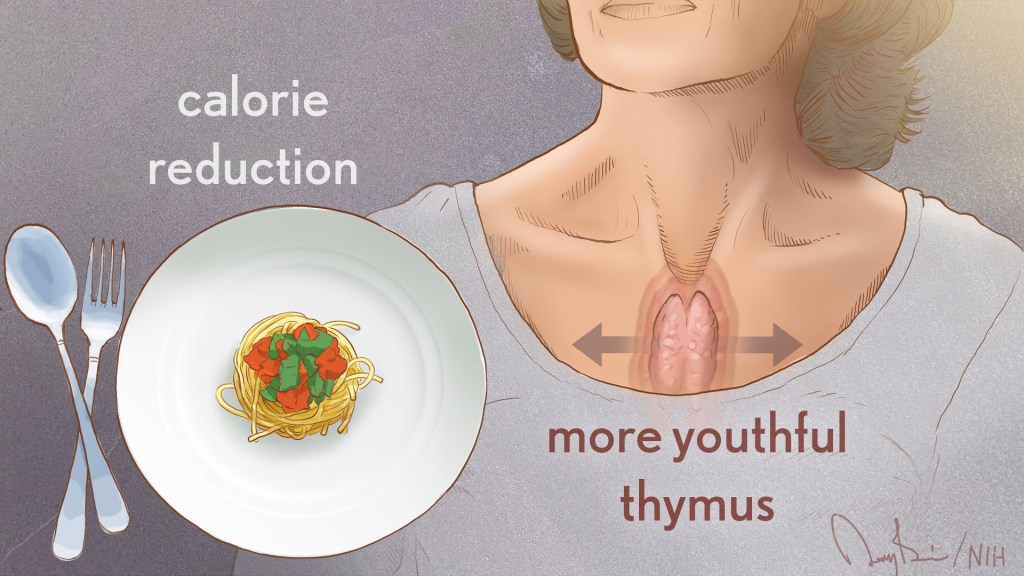
MRI
Visualizing The Placenta, a Critical but Poorly Understood Organ
Posted on by Diana W. Bianchi, M.D., Eunice Kennedy Shriver National Institute of Child Health and Human Development
The placenta is the Rodney Dangerfield of organs; it gets no respect, no respect at all. This short-lived but critical organ supports pregnancy by bringing nutrients and oxygen to the fetus, removing waste, providing immune protection, and producing hormones to support fetal development.
It also influences the lifelong health of both mother and child. Problems with the placenta can lead to preeclampsia, gestational diabetes, poor fetal growth, preterm birth, and stillbirth. Although we were all connected to one, the placenta is the least understood, and least studied, of all human organs.
What we do know about the human placenta largely comes from studying it after delivery. But that’s like studying the heart after it’s stopped beating. It doesn’t help us predict complications in time to avert a crisis.
To fill these knowledge gaps, NIH’s Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) developed the Human Placenta Project (HPP) to noninvasively study the placenta during pregnancy. Since 2014, this approximately $88 million collaborative research effort has been developing ultrasound, magnetic resonance imaging (MRI), and blood-based biomarker methods to study how the placenta functions in real time and in greater detail.
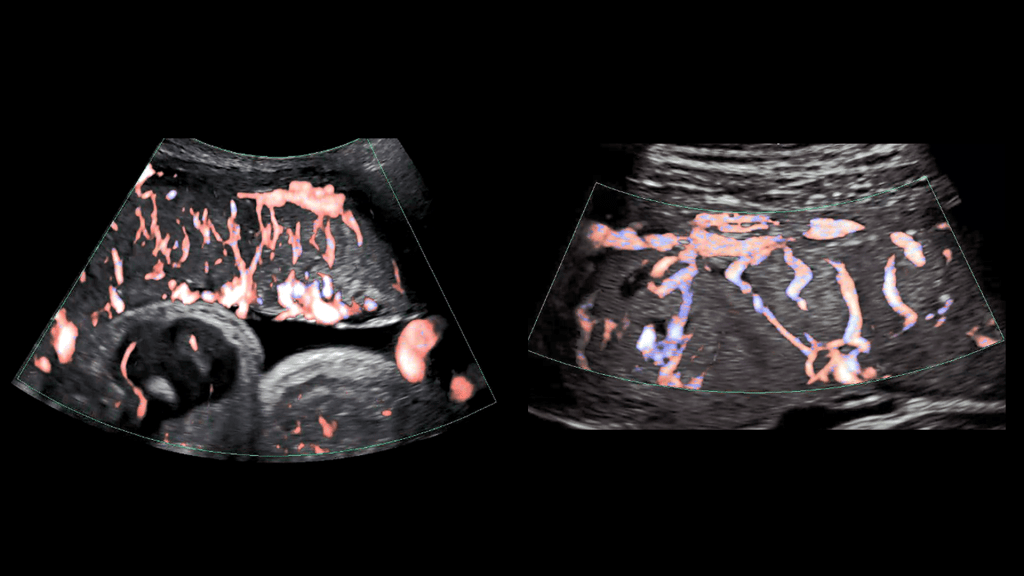
As illustrated in the image above, advanced ultrasound tools allowed HPP researchers at Eastern Virginia Medical School, Norfolk, and the University of Texas Medical Branch, Galveston, to gain a detailed look at the placenta’s intricate arrangement of blood vessels, or vasculature. By evaluating both fetal (left panel) and maternal (right panel) placental vasculature in 610 pregnant people starting at 13 weeks of gestation, the investigators aimed to identify early changes that predicted later complications.
They observed that such changes can start in the first trimester and affect both the vasculature and placental tissue. While further research is needed, these findings suggest that placental ultrasound monitoring can inform efforts to prevent and treat pregnancy complications.
Another HPP team led by Boston Children’s Hospital is developing an MRI strategy to monitor blood flow and oxygen transport through the placenta during pregnancy. Interpreting and visualizing MRI data of the placenta is challenging because of its variable shape, the tendency of muscles in the uterus to begin tightening or contracting well before labor [1], and other factors.
As shown in the video above, the researchers developed a way to account for the motion of the uterus and “freeze” the placenta to make it easier to study (left two panels of video) [2]. They also developed algorithms to better visualize the complex patterns of placental oxygen content during contractions (center panel) [3]. The scientists then carried out initial visualizations of blood flow through the placenta shortly after delivery (second panel from right) [4].
They now intend to map these MRI findings to the placenta itself after delivery (far right panel), which will allow them to explore how additional factors such as gene expression patterns and genetic variants contribute to placental function. Ultimately, they plan to apply these MRI techniques to monitor the placenta in real time during pregnancy and identify changes that indicate compromised function early enough to adjust maternal management as needed.
Other HPP efforts focus on identifying components in maternal blood that reflect the status of the placenta. For example, an HPP research team led by scientists at the University of California, Los Angeles, adapted non-invasive prenatal testing methods to analyze genetic material shed from the placenta into the maternal bloodstream. Their findings suggest that distinctive patterns in this genetic material detected early in pregnancy may indicate risk for later complications [5].
Another HPP team, led by investigators at Columbia University, New York, helped establish that extracellular RNAs (exRNAs) released by the placenta into maternal circulation reflect the placenta’s status at a cellular level beginning in the first trimester. To harness the potential of exRNA biomarkers, the investigators are optimizing methods to isolate, sequence, and analyze exRNAs in maternal blood.
These are just a few examples of the cutting-edge work being funded through the HPP, which complements NICHD’s longstanding investment in basic research to unravel the physiology of and real-time gene expression in the placenta. Unlocking the secrets of the placenta may one day help us to prevent and treat a range of common pregnancy complications, while also providing insights into other areas of science and medicine such as cardiovascular disease and aging. NICHD is committed to giving this important organ the respect it deserves.
References:
[1] Placental MRI: Effect of maternal position and uterine contractions on placental BOLD MRI measurements. Abaci Turk E, Abulnaga SM, Luo J, Stout JN, Feldman H, Turk A, Gagoski B, Wald LL, Adalsteinsson E, Roberts DJ, Bibbo C, Robinson JN, Golland P, Grant PE, Barth, Jr WH. Placenta. 2020 Jun 1; 95: 69-77.
[2] Spatiotemporal alignment of in utero BOLD-MRI series. Turk EA, Luo J, Gagoski B, Pascau J, Bibbo C, Robinson JN, Grant PE, Adalsteinsson E, Golland P, Malpica N. J Magn Reson Imaging. 2017 Aug;46(2):403-412.
[3] Volumetric parameterization of the placenta to a flattened template. Abulnaga SM, Turk EA, Bessmeltsev M, Grant PE, Solomon J, Golland P. IEEE transactions on medical imaging. 2022 April;41(4):925-936.
[4] Placental MRI: development of an MRI compatible ex vivo system for whole placenta dual perfusion. Stout JN, Rouhani S, Turk EA, Ha CG, Luo J, Rich K, Wald LL, Adalsteinsson E, Barth, Jr WH, Grant PE, Roberts DJ. Placenta. 2020 Nov 1; 101: 4-12.
[5] Cell-free DNA methylation and transcriptomic signature prediction of pregnancies with adverse outcomes. Del Vecchio G, Li Q, Li W, Thamotharan S, Tosevska A, Morselli M, Sung K, Janzen C, Zhou X, Pellegrini M, Devaskar SU. Epigenetics. 2021 Jun;16(6):642-661.
Links:
Human Placenta Project (Eunice Kennedy Shriver National Institute of Child Health and Human Development/NIH)
Preeclampsia (NICHD)
Understanding Gestational Diabetes (NICHD)
Preterm Labor and Birth (NICHD)
Stillbirth (NICHD)
Abuhamad Project Information (NIH RePORTER)
Grant Project Information (NIH RePORTER)
Devaskar Project Information (NIH RePORTER)
Williams Project Information (NIH RePORTER)
Note: Acting NIH Director Lawrence Tabak has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 10th in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
The Amazing Brain: A Sharper Image of the Pyramidal Tract
Posted on by Dr. Francis Collins
Flip the image above upside down, and the shape may remind you of something. If you think it resembles a pyramid, then you and a lot of great neuroscientists are thinking alike. What you are viewing is a colorized, 3D reconstruction of a pyramidal tract, which are bundles of nerve fibers that originate from the brain’s cerebral cortex and relay signals to the brainstem or the spinal cord. These signals control many important activities, including the voluntary movement of our arms, legs, head, and face.
For a while now, it’s been possible to combine a specialized form of magnetic resonance imaging (MRI) with computer modeling tools to produce 3D reconstructions of complicated networks of nerve fibers, such as the pyramidal tract. Still, for technical reasons, the quality of these reconstructions has remained poor in parts of the brain where nerve fibers cross at angles of 40 degrees or less.
The video above demonstrates how adding a sophisticated algorithm, called Orientation Distribution Function (ODF)-Fingerprinting, to such modeling can help overcome this problem when reconstructing a pyramidal tract. It has potential to enhance the reliability of these 3D reconstructions as neurosurgeons begin to use them to plan out their surgeries to help ensure they are carried out with the utmost safety and precision.
In the first second of the video, you see gray, fuzzy images from a diffusion MRI of the pyramidal tract. But, very quickly, a more colorful, detailed 3D reconstruction begins to appear, swiftly filling in from the top down. Colors are used to indicate the primary orientations of the nerve fibers: left to right (red), back to front (green), and top to bottom (blue). The orange, magenta, and other colors represent combinations of these primary directional orientations.
About three seconds into the video, a rough draft of the 3D reconstruction is complete. The top of the pyramidal tract looks pretty good. However, looking lower down, you can see distortions in color and relatively poor resolution of the nerve fibers in the middle of the tract—exactly where the fibers cross each other at angles of less than 40 degrees. So, researchers tapped into the power of their new ODF-Fingerprinting software to improve the image—and, starting about nine seconds into the video, you can see an impressive final result.
The researchers who produced this amazing video are Patryk Filipiak and colleagues in the NIH-supported lab of Steven Baete, Center for Advanced Imaging Innovation and Research, New York University Grossman School of Medicine, New York. The work paired diffusion MRI data from the NIH Human Connectome Project with the ODF-Fingerprinting algorithm, which was created by Baete to incorporate additional MRI imaging data on the shape of nerve fibers to infer their directionality [1].
This innovative approach to imaging recently earned Baete’s team second place in the 2021 “Show Us Your BRAINs” Photo and Video contest, sponsored by the NIH-led Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative. But researchers aren’t stopping there! They are continuing to refine ODF-Fingerprinting, with the aim of modeling the pyramidal tract in even higher resolution for use in devising new and better ways of helping people undergoing neurosurgery.
Reference:
[1] Fingerprinting Orientation Distribution Functions in diffusion MRI detects smaller crossing angles. Baete SH, Cloos MA, Lin YC, Placantonakis DG, Shepherd T, Boada FE. Neuroimage. 2019 Sep;198:231-241.
Links:
Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative (NIH)
Human Connectome Project (University of Southern California, Los Angeles)
Steven Baete (Center for Advanced Imaging Innovation and Research, New York University Grossman School of Medicine, New York)
Show Us Your BRAINs! Photo and Video Contest (BRAIN Initiative/NIH)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Neurological Disorders and Stroke; National Cancer Institute
COVID-19 Can Damage Hearts of Some College Athletes
Posted on by Dr. Francis Collins

There’s been quite a bit of discussion in the news lately about whether to pause or resume college athletics during the pandemic. One of the sticking points has been uncertainty about how to monitor the health of student athletes who test positive for SARS-CoV-2, the novel coronavirus that causes COVID-19. As a result, college medical staff don’t always know when to tell athletes that they’ve fully recovered and it’s safe to start training again.
The lack of evidence owes to two factors. Though it may not seem like it, this terrible coronavirus has been around for less than a year, and that’s provided little time to conduct the needed studies with young student athletes. But that’s starting to change. An interesting new study in the journal JAMA Cardiology provides valuable and rather worrisome early data from COVID-positive student athletes evaluated for an inflammation of the heart called myocarditis, a well-known complication [1].
Saurabh Rajpal and his colleagues at the Ohio State University, Columbus, used cardiac magnetic resonance imaging (MRI) to visualize the hearts of 26 male and female student athletes. They participated in a range of sports, including football, soccer, lacrosse, basketball, and track. All of the athletes were referred to the university’s sports medicine clinic this past summer after testing positive for SARS-CoV-2. All had mild or asymptomatic cases of COVID-19.
Even so, the MRI scans, taken 11-53 days after completion of quarantine, showed four of the student athletes (all males) had swelling and tissue damage to their hearts consistent with myocarditis. Although myocarditis often resolves on its own over time, severe cases can compromise the heart muscle’s ability to beat. That can lead to heart failure, abnormal heart rhythms, and even sudden death in competitive athletes with normal heart function [2].
The investigators also looked for more subtle findings of cardiac injury in these athletes, using a contrast agent called gadolinium and measuring its time to appear in the cardiac muscle during the study. Eight of the 26 athletes (31 percent) had late gadolinium enhancement, suggestive of prior myocardial injury.
Even though it’s a small study, these results certainly raise concerns. They add more evidence to a prior study, published by a German group, that suggested subtle cardiac consequences of SARS-CoV-2 infection may be common in adults [3].
Rajpal and his colleagues will continue to follow the athletes in their study for several more months. The researchers will keep an eye out for other lingering symptoms of COVID-19, generate more cardiac MRI data, and perform exercise testing.
As this study shows, we still have a lot to learn about the long-term consequences of COVID-19, which can take people on different paths to recovery. For athletes, that path is the challenge to return to top physical shape and feel ready to compete at a high level. But getting back in uniform must also be done safely to minimize any risks to an athlete’s long-term health and wellbeing. The more science-based evidence that’s available, the more prepared athletes at large and small colleges will be to compete safely in this challenging time.
References:
[1] Cardiovascular magnetic resonance findings in competitive athletes recovering from COVID-19 infection. Rajpal S, Tong MS, Borchers J, et al. JAMA Cardiol. 2020 September 11. [Published online ahead of print.]
[2] Eligibility and disqualification recommendations for competitive athletes with cardiovascular abnormalities: Task Force 3: Hypertrophic cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy and other cardiomyopathies, and myocarditis. Maron BJ, Udelson JE, Bonow RO, et al. Circulation. 2015;132(22):e273-e280.
[3] Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from Coronavirus Disease 2019 (COVID-19). Puntmann VO, Carej ML, Wieters I. JAMA Cardiol. 2020 Jul 27:e203557. [Published online ahead of print.]
Links:
Coronavirus (COVID-19) (NIH)
Heart Inflammation (National Heart, Lung, and Blood Institute/NIH)
Saurabh Rajpal (Ohio State College of Medicine, Columbus)
Prostate Cancer: Combined Biopsy Strategy Makes for More Accurate Diagnosis
Posted on by Dr. Francis Collins

Last year, nearly 175,000 American men were diagnosed with prostate cancer [1]. Most got the bad news after a blood test or physical exam raised concerns that warranted a biopsy of the prostate, a walnut-sized gland just below the bladder.
Traditional biopsies sample tissue from 12 systematically placed points within the prostate that are blind to tumor locations. Such procedures have helped to save many lives, but are prone to missing or misclassifying prostate cancers, which has led doctors both to over and under treat their patients.
Now, there may be a better approach. In a study of more than 2,000 men, NIH researchers and their colleagues recently found that combining the 12-point biopsy with magnetic resonance imaging (MRI)-targeted biopsy during the same session more accurately diagnoses prostate cancer than either technique alone [2].
The findings address a long-standing challenge in prostate cancer diagnostics: performing a thorough prostate biopsy to allow a pathologist to characterize as accurately as possible the behavior of a tumor. Some prostate tumors are small, slow growing, and can be monitored closely without treatment. Other tumors are aggressive and can grow rapidly, requiring immediate intervention with hormonal therapy, radiation, or surgery.
But performing a thorough prostate biopsy can run into technical difficulties. The 12-point biopsy blindly samples tissue from across the prostate gland, but it can miss a cancer by not probing in the right places.
Several years ago, researchers at the NIH Clinical Center, Bethesda, MD, envisioned a solution. They’d use specially designed MRI images of a man’s prostate to guide the biopsy needle to areas in the prostate that look suspicious and deserve a closer look under a microscope.
Through a cooperative research-and-development agreement, NIH and the now- Florida-based Philips Healthcare created an office-based, outpatient prostate biopsy device, called UroNav, that was later approved by the Food and Drug Administration. The UroNav system relies on software that overlays MRI images highlighting suspicious areas onto real-time ultrasound images of the prostate that are traditionally used to guide biopsy procedures.
The new technology led to a large clinical study led by Peter Pinto, a researcher with NIH’s National Cancer Institute. The study results, published in 2015, found that the MRI-targeted approach was indeed superior to the 12-point biopsy at detecting aggressive prostate cancers [3].
But some doctors had questions about how best to implement the UroNav system and whether it could replace the 12-point biopsy. The uncertainty led to a second clinical study to nail down more answers, and the results were just published in The New England Journal of Medicine.
The research team enrolled 2,103 men who had visible prostate abnormalities on an MRI. Once in the study, each man underwent both the 12-point blind biopsy and the MRI-targeted approach—all in a single office visit. Based on this two-step approach, 1,312 people were diagnosed with prostate cancer. Of that total, 404 men had evidence of aggressive cancer, and had their prostates surgically removed.
The researchers then compared the diagnoses from each approach alone versus the two combined. The data showed that the combined biopsy found 208 cancers that the standard 12-point biopsy alone would have missed. Adding the MRI-targeted biopsy also helped doctors find and sample the more aggressive cancers. This allowed them to upgrade the diagnosis of 458 cancers to aggressive and in need of more full treatment.
Combining the two approaches also led to more accurate diagnoses. By carefully analyzing the 404 removed prostates and comparing them to the biopsy results, the researchers found the 12-point biopsy missed the most aggressive cancers about 40 percent of the time. But the MRI-targeted approach alone missed it about 30 percent of the time. Combined, they did much better, underestimating the severity of less than 15 percent of the cancers.
Even better, the combined biopsy missed only 3.5 percent of the most aggressive tumors. That’s compared to misses of about 17 percent for the most-aggressive cancers for the 12-point biopsy alone and about 9 percent for MRI-targeted biopsy alone.
It may take time for doctors to change how they detect prostate cancer in their practices. But the findings show that combining both approaches will significantly improve the accuracy of diagnosing prostate cancer. This will, in turn, help to reduce risk of suboptimal treatment (too much or too little) by allowing doctors and patients to feel more confident in the biopsy results. That should come as good news now and in the future for the families and friends of men who will need an accurate prostate biopsy to make informed treatment decisions.
References:
[1] Cancer State Facts: Prostate Cancer. National Cancer Institute Surveillance, Epidemiology, and End Results Program.
[2] MRI-targeted, systematic, and combined biopsy for prostate cancer diagnosis. Ahdoot M, Wilbur AR, Reese SE, Lebastchi AH, Mehralivand S, Gomella PT, Bloom J, Gurram S, Siddiqui M, Pinsky P, Parnes H, Linehan WM, Merino M, Choyke PL, Shih JH, Turkbey B, Wood BJ, Pinto PA. N Engl J Med. 2020 Mar 5;382(10):917-928.
[3] Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. Siddiqui M, Rais-Bahrami, George AK, Rothwax J, Shakir N, Okoro C, Raskolnikov D, Parnes HL, Linehan WM, Merino MJ, Simon RM, Choyke PL, Wood BJ, and Pinto PA. JAMA. 2015 January 27;313(4):390-397.
Links:
Prostate Cancer (National Cancer Institute/NIH)
Video: MRI-Targeted Prostate Biopsy (YouTube)
Pinto Lab (National Cancer Institute/NIH)
NIH Support: National Cancer Institute; NIH Clinical Center
The Amazing Brain: Mapping Brain Circuits in Vivid Color
Posted on by Dr. Francis Collins
Hop aboard as we fly up, down, left, and right through the information highways of the human brain! This captivating and eye-catching video was one of the winners of the 2019 “Show us Your Brain!” contest sponsored by the NIH-led Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative.
The video travels through several portions of the brain’s white matter—bundles of fiber that carry nerve signals between the brain and the body, as well as within the brain itself. Fiber colors indicate directionality: left-right fibers (red), front-back fibers (green), and top-bottom fibers (blue).
Looking from the back, we start our journey deep within the brain in the limbic system, the area that helps control emotion, learning, and memory. About three seconds in, visual fibers pop into view extending from the eyes to various brain areas into the occipital lobe (one of four major brain lobes) in the back of the brain.
About two seconds later, flying over top as the brain starts rotating, we see various fiber bundles spray upward throughout the cerebral cortex, communicating information related to language processing, short-term memory, and other functions. About halfway through the video, several green bundles emerge arching across the brain’s midline. These bundles, called the corpus callosum, house the fibers enabling communication between left and right sides of the brain. Finally, the video closes as we see many different fiber bundles lighting up all over, enabling communication between different cortical and subcortical portions of the brain through association and projection pathways.
Dynamic maps like these are created using a 3D imaging technique called diffusion MRI tractography [1]. The technique tracks subtle pathways of water movement in the brain, and allows researchers to model the physical properties (connectional anatomy) that underlie the brain’s electrical properties (neuronal signaling). Postdoctoral researcher Ryan Cabeen and Arthur Toga, director of the University of Southern California Mark and Mary Stevens Neuroimaging and Informatics Institute, Los Angeles, used the method to study how white matter changes in developing and aging brains, as well as in brains affected by neurodegenerative or neurological disorders.
Scientific animator Jim Stanis produced the video with Cabeen and Toga. The team first created a population-averaged brain using high-quality diffusion MRI datasets from the Human Connectome Project ,and then used sophisticated computational tools to delineate each bundle manually .
The tractography technique lets scientists visualize and quantitatively analyze the brain’s wiring patterns, complementing our understanding of how the brain functions. Such methods are especially useful to learn about the organization of deep-brain areas that remain out of reach for scientists using current tools and imaging techniques.
Reference:
[1] Kernel regression estimation of fiber orientation mixtures in diffusion MRI. Cabeen RP, Bastin ME, Laidlaw DH. Neuroimage. 2016 Feb 15;127:158-172.
Links:
Arthur Toga (USC Mark and Mary Stevens Neuroimaging and Informatics Institute, Los Angeles)
Ryan Cabeen (USC Mark and Mary Stevens Neuroimaging and Informatics Institute)
qitwiki—Information about the Quantitative Imaging Toolkit (USC)
Human Connectome Project (USC)
Show Us Your Brain Contest! (BRAIN Initiative/NIH)
Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative (NIH)
NIH Support: National Institute of Neurological Disorders and Stroke; National Institute of Mental Health
Brain in Motion
Posted on by Dr. Francis Collins
Credit: Itamar Terem, Stanford University, Palo Alto, CA, and Samantha Holdsworth, University of Auckland, New Zealand
Though our thoughts can wander one moment and race rapidly forward the next, the brain itself is often considered to be motionless inside the skull. But that’s actually not correct. When the heart beats, the pumping force reverberates throughout the body and gently pulsates the brain. What’s been tricky is capturing these pulsations with existing brain imaging technologies.
Recently, NIH-funded researchers developed a video-based approach to magnetic resonance imaging (MRI) that can record these subtle movements [1]. Their method, called phase-based amplified MRI (aMRI), magnifies those tiny movements, making them more visible and quantifiable. The latest aMRI method, developed by a team including Itamar Terem at Stanford University, Palo Alto, CA, and Mehmet Kurt at Stevens Institute of Technology, Hoboken, NJ. It builds upon an earlier method developed by Samantha Holdsworth at New Zealand’s University of Auckland and Stanford’s Mahdi Salmani Rahimi [2].
Seven More Awesome Technologies Made Possible by Your Tax Dollars
Posted on by Dr. Francis Collins
We live in a world energized by technological advances, from that new app on your smartphone to drones and self-driving cars. As you can see from this video, NIH-supported researchers are also major contributors, developing a wide range of amazing biomedical technologies that offer tremendous potential to improve our health.
Produced by the NIH’s National Institute of Biomedical Imaging and Bioengineering (NIBIB), this video starts by showcasing some cool fluorescent markers that are custom-designed to light up specific cells in the body. This technology is already helping surgeons see and remove tumor cells with greater precision in people with head and neck cancer [1]. Further down the road, it might also be used to light up nerves, which can be very difficult to see—and spare—during operations for cancer and other conditions.
Other great things to come include:
- A wearable tattoo that detects alcohol levels in perspiration and wirelessly transmits the information to a smartphone.
- Flexible coils that produce high quality images during magnetic resonance imaging (MRI) [2-3]. In the future, these individualized, screen-printed coils may improve the comfort and decrease the scan times of people undergoing MRI, especially infants and small children.
- A time-release capsule filled with a star-shaped polymer containing the anti-malarial drug ivermectin. The capsule slowly dissolves in the stomach over two weeks, with the goal of reducing the need for daily doses of ivermectin to prevent malaria infections in at-risk people [4].
- A new radiotracer to detect prostate cancer that has spread to other parts of the body. Early clinical trial results show the radiotracer, made up of carrier molecules bonded tightly to a radioactive atom, appears to be safe and effective [5].
- A new supercooling technique that promises to extend the time that organs donated for transplantation can remain viable outside the body [6-7]. For example, current technology can preserve donated livers outside the body for just 24 hours. In animal studies, this new technique quadruples that storage time to up to four days.
- A wearable skin patch with dissolvable microneedles capable of effectively delivering an influenza vaccine. This painless technology, which has produced promising early results in humans, may offer a simple, affordable alternative to needle-and-syringe immunization [8].
If you like what you see here, be sure to check out this previous NIH video that shows six more awesome biomedical technologies that your tax dollars are helping to create. So, let me extend a big thanks to you from those of us at NIH—and from all Americans who care about the future of their health—for your strong, continued support!
References:
[1] Image-guided surgery in cancer: A strategy to reduce incidence of positive surgical margins. Wiley Interdiscip Rev Syst Biol Med. 2018 Feb 23.
[2] Screen-printed flexible MRI receive coils. Corea JR, Flynn AM, Lechêne B, Scott G, Reed GD, Shin PJ, Lustig M, Arias AC. Nat Commun. 2016 Mar 10;7:10839.
[3] Printed Receive Coils with High Acoustic Transparency for Magnetic Resonance Guided Focused Ultrasound. Corea J, Ye P, Seo D, Butts-Pauly K, Arias AC, Lustig M. Sci Rep. 2018 Feb 21;8(1):3392.
[4] Oral, ultra-long-lasting drug delivery: Application toward malaria elimination goals. Bellinger AM, Jafari M1, Grant TM, Zhang S, Slater HC, Wenger EA, Mo S, Lee YL, Mazdiyasni H, Kogan L, Barman R, Cleveland C, Booth L, Bensel T, Minahan D, Hurowitz HM, Tai T, Daily J, Nikolic B, Wood L, Eckhoff PA, Langer R, Traverso G. Sci Transl Med. 2016 Nov 16;8(365):365ra157.
[5] Clinical Translation of a Dual Integrin avb3– and Gastrin-Releasing Peptide Receptor–Targeting PET Radiotracer, 68Ga-BBN-RGD. Zhang J, Niu G, Lang L, Li F, Fan X, Yan X, Yao S, Yan W, Huo L, Chen L, Li Z, Zhu Z, Chen X. J Nucl Med. 2017 Feb;58(2):228-234.
[6] Supercooling enables long-term transplantation survival following 4 days of liver preservation. Berendsen TA, Bruinsma BG, Puts CF, Saeidi N, Usta OB, Uygun BE, Izamis ML, Toner M, Yarmush ML, Uygun K. Nat Med. 2014 Jul;20(7):790-793.
[7] The promise of organ and tissue preservation to transform medicine. Giwa S, Lewis JK, Alvarez L, Langer R, Roth AE, et a. Nat Biotechnol. 2017 Jun 7;35(6):530-542.
[8] The safety, immunogenicity, and acceptability of inactivated influenza vaccine delivered by microneedle patch (TIV-MNP 2015): a randomised, partly blinded, placebo-controlled, phase 1 trial. Rouphael NG, Paine M, Mosley R, Henry S, McAllister DV, Kalluri H, Pewin W, Frew PM, Yu T, Thornburg NJ, Kabbani S, Lai L, Vassilieva EV, Skountzou I, Compans RW, Mulligan MJ, Prausnitz MR; TIV-MNP 2015 Study Group.
Links:
National Institute of Biomedical Imaging and Bioengineering (NIH)
Center for Wearable Sensors (University of California, San Diego)
Hyperpolarized MRI Technology Resource Center (University of California, San Francisco)
Center for Engineering in Medicine (Massachusetts General Hospital, Boston)
Center for Drug Design, Development and Delivery (Georgia Tech University, Atlanta)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of Allergy and Infectious Diseases
New Imaging Approach Reveals Lymph System in Brain
Posted on by Dr. Francis Collins
Considering all the recent advances in mapping the complex circuitry of the human brain, you’d think we’d know all there is to know about the brain’s basic anatomy. That’s what makes the finding that I’m about to share with you so remarkable. Contrary to what I learned in medical school, the body’s lymphatic system extends to the brain—a discovery that could revolutionize our understanding of many brain disorders, from Alzheimer’s disease to multiple sclerosis (MS).
Researchers from the National Institute of Neurological Disorders and Stroke (NINDS), the National Cancer Institute (NCI), and the University of Virginia, Charlottesville made this discovery by using a special MRI technique to scan the brains of healthy human volunteers [1]. As you see in this 3D video created from scans of a 47-year-old woman, the brain—just like the neck, chest, limbs, and other parts of the body—possesses a network of lymphatic vessels (green) that serves as a highway to circulate key immune cells and return metabolic waste products to the bloodstream.
Brain Scans Show Early Signs of Autism Spectrum Disorder
Posted on by Dr. Francis Collins

Source: Getty Images
For children with autism spectrum disorder (ASD), early diagnosis is critical to allow for possible interventions at a time when the brain is most amenable to change. But that’s been tough to implement for a simple reason: the symptoms of ASD, such as communication difficulties, social deficits, and repetitive behaviors, often do not show up until a child turns 2 or even 3 years old.
Now, an NIH-funded research team has news that may pave the way for earlier detection of ASD. The key is to shift the diagnostic focus from how kids act to how their brains grow. In their brain imaging study, the researchers found that, compared to other children, youngsters with ASD showed unusually rapid brain growth from infancy to age 2. In fact, the growth differences were already evident by their first birthdays, well before autistic behaviors typically emerge.
Next Page