head and neck cancer
Seven More Awesome Technologies Made Possible by Your Tax Dollars
Posted on by Dr. Francis Collins
We live in a world energized by technological advances, from that new app on your smartphone to drones and self-driving cars. As you can see from this video, NIH-supported researchers are also major contributors, developing a wide range of amazing biomedical technologies that offer tremendous potential to improve our health.
Produced by the NIH’s National Institute of Biomedical Imaging and Bioengineering (NIBIB), this video starts by showcasing some cool fluorescent markers that are custom-designed to light up specific cells in the body. This technology is already helping surgeons see and remove tumor cells with greater precision in people with head and neck cancer [1]. Further down the road, it might also be used to light up nerves, which can be very difficult to see—and spare—during operations for cancer and other conditions.
Other great things to come include:
- A wearable tattoo that detects alcohol levels in perspiration and wirelessly transmits the information to a smartphone.
- Flexible coils that produce high quality images during magnetic resonance imaging (MRI) [2-3]. In the future, these individualized, screen-printed coils may improve the comfort and decrease the scan times of people undergoing MRI, especially infants and small children.
- A time-release capsule filled with a star-shaped polymer containing the anti-malarial drug ivermectin. The capsule slowly dissolves in the stomach over two weeks, with the goal of reducing the need for daily doses of ivermectin to prevent malaria infections in at-risk people [4].
- A new radiotracer to detect prostate cancer that has spread to other parts of the body. Early clinical trial results show the radiotracer, made up of carrier molecules bonded tightly to a radioactive atom, appears to be safe and effective [5].
- A new supercooling technique that promises to extend the time that organs donated for transplantation can remain viable outside the body [6-7]. For example, current technology can preserve donated livers outside the body for just 24 hours. In animal studies, this new technique quadruples that storage time to up to four days.
- A wearable skin patch with dissolvable microneedles capable of effectively delivering an influenza vaccine. This painless technology, which has produced promising early results in humans, may offer a simple, affordable alternative to needle-and-syringe immunization [8].
If you like what you see here, be sure to check out this previous NIH video that shows six more awesome biomedical technologies that your tax dollars are helping to create. So, let me extend a big thanks to you from those of us at NIH—and from all Americans who care about the future of their health—for your strong, continued support!
References:
[1] Image-guided surgery in cancer: A strategy to reduce incidence of positive surgical margins. Wiley Interdiscip Rev Syst Biol Med. 2018 Feb 23.
[2] Screen-printed flexible MRI receive coils. Corea JR, Flynn AM, Lechêne B, Scott G, Reed GD, Shin PJ, Lustig M, Arias AC. Nat Commun. 2016 Mar 10;7:10839.
[3] Printed Receive Coils with High Acoustic Transparency for Magnetic Resonance Guided Focused Ultrasound. Corea J, Ye P, Seo D, Butts-Pauly K, Arias AC, Lustig M. Sci Rep. 2018 Feb 21;8(1):3392.
[4] Oral, ultra-long-lasting drug delivery: Application toward malaria elimination goals. Bellinger AM, Jafari M1, Grant TM, Zhang S, Slater HC, Wenger EA, Mo S, Lee YL, Mazdiyasni H, Kogan L, Barman R, Cleveland C, Booth L, Bensel T, Minahan D, Hurowitz HM, Tai T, Daily J, Nikolic B, Wood L, Eckhoff PA, Langer R, Traverso G. Sci Transl Med. 2016 Nov 16;8(365):365ra157.
[5] Clinical Translation of a Dual Integrin avb3– and Gastrin-Releasing Peptide Receptor–Targeting PET Radiotracer, 68Ga-BBN-RGD. Zhang J, Niu G, Lang L, Li F, Fan X, Yan X, Yao S, Yan W, Huo L, Chen L, Li Z, Zhu Z, Chen X. J Nucl Med. 2017 Feb;58(2):228-234.
[6] Supercooling enables long-term transplantation survival following 4 days of liver preservation. Berendsen TA, Bruinsma BG, Puts CF, Saeidi N, Usta OB, Uygun BE, Izamis ML, Toner M, Yarmush ML, Uygun K. Nat Med. 2014 Jul;20(7):790-793.
[7] The promise of organ and tissue preservation to transform medicine. Giwa S, Lewis JK, Alvarez L, Langer R, Roth AE, et a. Nat Biotechnol. 2017 Jun 7;35(6):530-542.
[8] The safety, immunogenicity, and acceptability of inactivated influenza vaccine delivered by microneedle patch (TIV-MNP 2015): a randomised, partly blinded, placebo-controlled, phase 1 trial. Rouphael NG, Paine M, Mosley R, Henry S, McAllister DV, Kalluri H, Pewin W, Frew PM, Yu T, Thornburg NJ, Kabbani S, Lai L, Vassilieva EV, Skountzou I, Compans RW, Mulligan MJ, Prausnitz MR; TIV-MNP 2015 Study Group.
Links:
National Institute of Biomedical Imaging and Bioengineering (NIH)
Center for Wearable Sensors (University of California, San Diego)
Hyperpolarized MRI Technology Resource Center (University of California, San Francisco)
Center for Engineering in Medicine (Massachusetts General Hospital, Boston)
Center for Drug Design, Development and Delivery (Georgia Tech University, Atlanta)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of Allergy and Infectious Diseases
The Science of Saliva
Posted on by Dr. Francis Collins
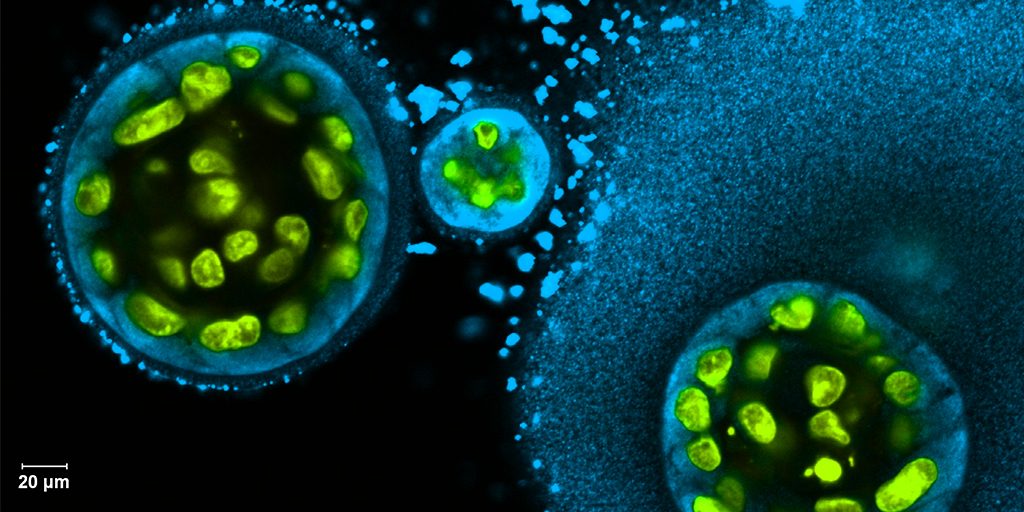
Credit: Swati Pradhan-Bhatt, Christiana Care Health System, Newark, DE
Whether it’s salmon sizzling on the grill or pizza fresh from the oven, you probably have a favorite food that makes your mouth water. But what if your mouth couldn’t water—couldn’t make enough saliva? When salivary glands stop working and the mouth becomes dry, either from disease or as a side effect of medical treatment, the once-routine act of eating can become a major challenge.
To help such people, researchers are now trying to engineer replacement salivary glands. While the research is still in the early stages, this image captures a crucial first step in the process: generating 3D structures of saliva-secreting cells (yellow). When grown on a scaffold of biocompatible polymers infused with factors to encourage development, these cells cluster into spherical structures similar to those seen in salivary glands. And they don’t just look like salivary cells, they act like them, producing the distinctive enzyme in saliva, alpha amylase (blue).
Optimizing Radio-Immunotherapy for Cancer
Posted on by Dr. Francis Collins

Zachary Morris
Credit: Alan Leon
Zachary Morris has certainly done some memorable things. As a Rhodes Scholar, he once attended an evening reception at Buckingham Palace, played a game of pick-up football with former President Bill Clinton, and traveled to South Africa to take a Robben Island Prison tour, led by the late Nelson Mandela. But something the young radiation oncologist did during his medical residency could prove even more momentous. He received a special opportunity from the American Board of Radiology to join others in studying how to pair radiation therapy with the emerging cancer treatment strategy of immunotherapy.
Morris’s studies in animals showed that the two treatments have a unique synergy, generating a sustained tumor-specific immune response that’s more potent than either therapy alone. But getting this combination therapy just right to optimize its cancer-fighting abilities remains complicated. Morris, now a researcher and clinician at the University of Wisconsin School of Medicine and Public Health, Madison, has received a 2017 NIH Director’s Early Independence Award to look deeper into this promising approach. He and his collaborators will use what they learn to better inform their future early stage clinical trials of radio-immunotherapy starting with melanoma, head and neck cancers, and neuroblastoma.
Head and Neck Cancer: Building the Evidence Base for Precision Oncology
Posted on by Dr. Francis Collins

Caption: Triple immunohistochemical stained oral squamous cell carcinoma: nuclei in brown, cytoplasm in red, and cytoplasmic membranes in blue green.
Credit: Alfredo A. Molinolo, Oral and Pharyngeal Cancer Branch, National Institute of Dental and Craniofacial Research, NIH
An exciting new era in cancer research is emerging, called precision oncology. It builds on decades of research establishing that cancers start with glitches in the genome, the cell’s instruction book. Researchers have now identified numerous ways that mutations in susceptible genes can drive the cancer process. Knowing where and how to look for them brings greater precision to diagnosing cancers and gives doctors key clues about which treatments might work and which ones won’t.
To build a firmer evidence base for precision oncology, more and more cancer genomes, from many different body sites, must be analyzed for clues about the drivers of the malignant process. That’s why it’s always exciting to see a new genomic analysis that adds substantially to our understanding of a common tumor. The latest to appear, published online at the journal Nature, comes from an NIH-supported study on the most common type of head and neck cancer, called squamous cell carcinoma. The technologically advanced analysis confirms that many previously suspected genes do indeed play a role in head and neck cancer. But that’s not all. The new data also identify several previously unknown subtypes of this cancer. The first descriptions of the abnormal molecular wiring in these subtypes are outlined, suggesting possible strategies to neutralize or destroy the cancer cells. That’s potentially good news to help guide and inform the treatment of the estimated 55,000 Americans who are diagnosed with a head and neck cancer each year.
Different Cancers Can Share Genetic Signatures
Posted on by Dr. Francis Collins
Cancer is a disease of the genome. It arises when genes involved in promoting or suppressing cell growth sustain mutations that disturb the normal stop and go signals. There are more than 100 different types of cancer, most of which derive their names and current treatment based on their tissue of origin—breast, colon, or brain, for example. But because of advances in DNA sequencing and analysis, that soon may be about to change.
Using data generated through The Cancer Genome Atlas, NIH-funded researchers recently compared the genomic fingerprints of tumor samples from nearly 3,300 patients with 12 types of cancer: acute myeloid leukemia, bladder, brain (glioblastoma multiforme), breast, colon, endometrial, head and neck, kidney, lung (adenocarcinoma and squamous cell carcinoma), ovarian, and rectal. Confirming but greatly extending what smaller studies have shown, the researchers discovered that even when cancers originate from vastly different tissues, they can show similar features at the DNA level