chromosome
The Perfect Cytoskeletal Storm
Posted on by Dr. Francis Collins
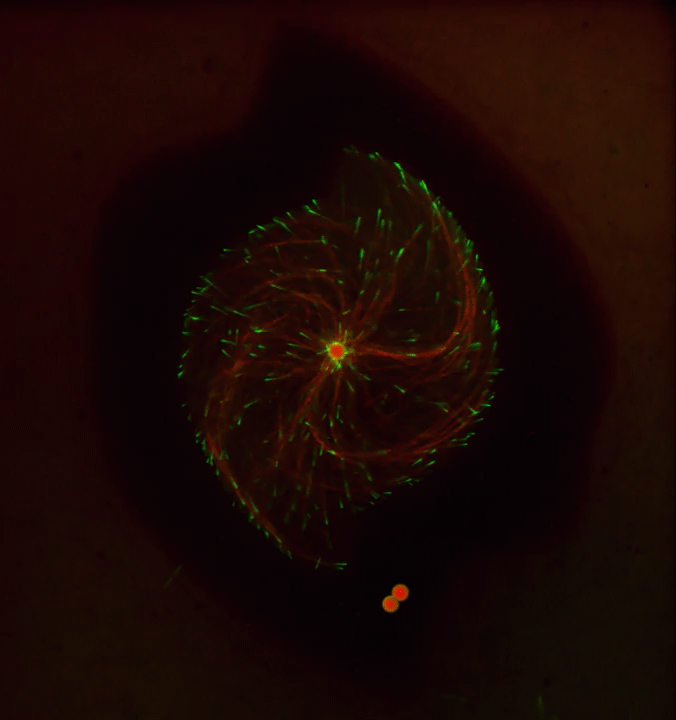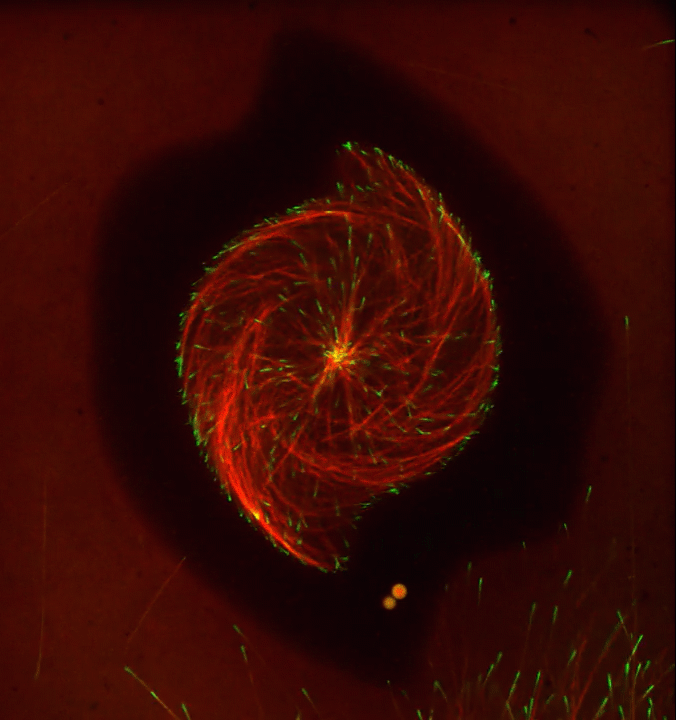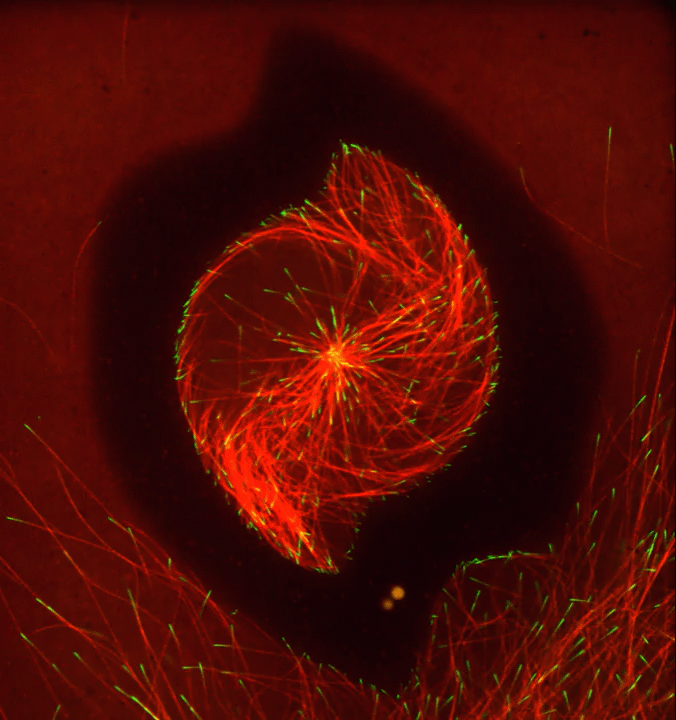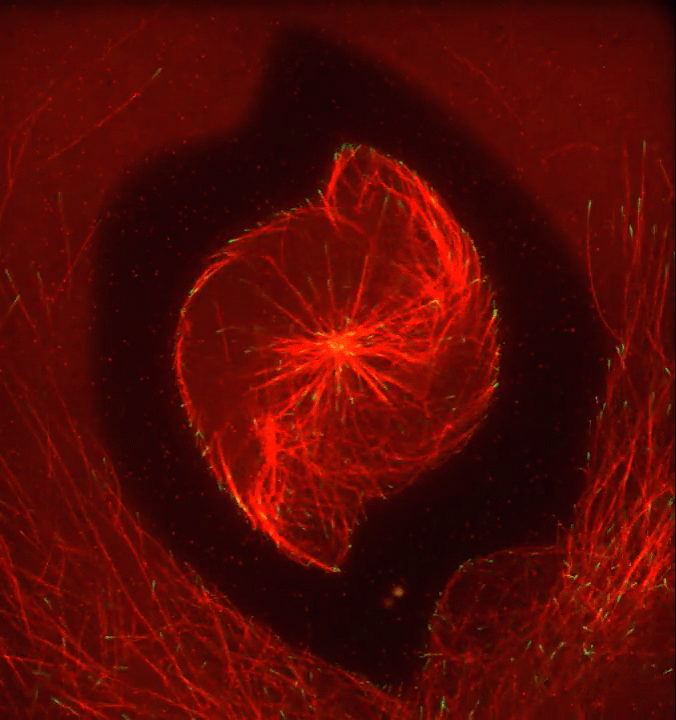
Ever thought about giving cell biology a whirl? If so, I suggest you sit down and take a look at this full-blown cytoskeletal “storm,” which provides a spectacular dynamic view of the choreography of life.
Before a cell divides, it undergoes a process called mitosis that copies its chromosomes and produces two identical nuclei. As part of this process, microtubules, which are structural proteins that help make up the cell’s cytoskeleton, reorganize the newly copied chromosomes into a dense, football-shaped spindle. The position of this mitotic spindle tells the cell where to divide, allowing each daughter cell to contain its own identical set of DNA.
To gain a more detailed view of microtubules in action, researchers designed an experimental system that utilizes an extract of cells from the African clawed frog (Xenopus laevis). As the video begins, a star-like array of microtubules (red) radiate outward in an apparent effort to prepare for cell division. In this configuration, the microtubules continually adjust their lengths with the help of the protein EB-1 (green) at their tips. As the microtubules grow and bump into the walls of a lab-generated, jelly-textured enclosure (dark outline), they buckle—and the whole array then whirls around the center.
Abdullah Bashar Sami, a Ph.D. student in the NIH-supported lab of Jesse “Jay” Gatlin, University of Wyoming, Laramie, shot this movie as a part his basic research to explore the still poorly understood physical forces generated by microtubules. The movie won first place in the 2019 Green Fluorescent Protein Image and Video Contest sponsored by the American Society for Cell Biology. The contest honors the 25th anniversary of the discovery of green fluorescent protein (GFP), which transformed cell biology and earned the 2008 Nobel Prize in Chemistry for three scientists who had been supported by NIH.
Like many movies, the setting was key to this video’s success. The video was shot inside a microfluidic chamber, designed in the Gatlin lab, to study the physics of microtubule assembly just before cells divide. The tiny chamber holds a liquid droplet filled with the cell extract.
When the liquid is exposed to an ultra-thin beam of light, it forms a jelly-textured wall, which traps the molecular contents inside [1]. Then, using time-lapse microscopy, the researchers watch the mechanical behavior of GFP-labeled microtubules [2] to see how they work to position the mitotic spindle. To do this, microtubules act like shapeshifters—scaling to adjust to differences in cell size and geometry.
The Gatlin lab is continuing to use their X. laevis system to ask fundamental questions about microtubule assembly. For many decades, both GFP and this amphibian model have provided cell biologists with important insights into the choreography of life, and, as this work shows, we can expect much more to come!
References:
[1] Microtubule growth rates are sensitive to global and local changes in microtubule plus-end density. Geisterfer ZM, Zhu D, Mitchison T, Oakey J, Gatlin JC. November 20, 2019.
[2] Tau-based fluorescent protein fusions to visualize microtubules. Mooney P, Sulerud T, Pelletier JF, Dilsaver MR, et al. Cytoskeleton (Hoboken). 2017 Jun;74(6):221-232.
Links:
Mitosis (National Human Genome Research Institute/NIH)
Gatlin Lab (University of Wyoming, Laramie)
Green Fluorescent Protein Image and Video Contest (American Society for Cell Biology, Bethesda, MD)
2008 Nobel Prize in Chemistry (Nobel Foundation, Stockholm, Sweden)
NIH Support: National Institute of General Medical Sciences
Finding New Genetic Mutations Amid Healthy Cells
Posted on by Dr. Francis Collins

You might recall learning in biology class that the cells constantly replicating and dividing in our bodies all carry the same DNA, inherited in equal parts from each parent. But it’s become increasingly clear in recent years that even seemingly healthy tissues contain neighborhoods of cells bearing their own acquired genetic mutations. The question is: What do all those altered cells mean for our health?
With support from a 2018 NIH Director’s New Innovator Award, Po-Ru Loh, Harvard Medical School, Boston, is on a quest to find out, though without the need for sequencing lots of DNA in his own lab. Loh will instead develop ultrasensitive computational tools to pick up on those often-subtle alterations within the vast troves of genomic data already stored in databases around the world.
How is that possible? The math behind it might be complex, but the underlying idea is surprisingly simple. His algorithms look for spots in the genome where a slight imbalance exists in the quantity of DNA inherited from mom versus dad.
Actually, Loh can’t tell from the data which parent provided any snippet of chromosomal DNA. But looking at DNA sequenced from a mixture of many cells, he can infer which stretches of DNA were most likely inherited together from a single parent.
Any slight skew in those quantities point the way to genomic territory where a tiny portion of chromosomal DNA either went missing or became duplicated in some cells. This common occurrence, especially in older adults, leads to a condition called genetic mosaicism, meaning that, contrary to most biology textbooks, all cells aren’t exactly the same.
By detecting those subtle imbalances in the data, Loh can pinpoint small DNA alterations, even when they occur in 1 in 1,000 cells collected from a person’s bloodstream, saliva, or tissues. That’s the kind of sensitivity that most scientists would not have thought possible.
Loh has already begun putting his new computational approach to work, as reported in Nature last year [1]. In DNA data from blood samples of more than 150,000 participants in the United Kingdom Biobank, his method uncovered well over 8,000 mosaic chromosomal alterations.
The study showed that some of those alterations were associated with an increased risk of developing blood cancers. However, it’s important to note that most people with evidence of mosaicism won’t go on to develop cancer. The researchers also made the unexpected discovery that some individuals carried genetic variants that made them more prone than others to pick up new mutations in their blood cells.
What’s especially exciting is Loh’s computational tools now make it possible to search for signs of mosaicism within all the genetic data that’s ever been generated. Even more importantly, these tools will allow Loh and other researchers to ask and answer important questions about the consequences of mosaicism for a wide range of diseases.
Reference:
[1] Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Loh PR, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, Birmann BM, Talkowski ME, Bakhoum SF, McCarroll SA, Price AL. Nature. 2018 Jul;559(7714):350-355.
Links:
Loh Lab (Harvard Medical School, Boston)
Loh Project Information (NIH RePORTER)
NIH Director’s New Innovator Award (Common Fund)
NIH Support: Common Fund; National Institute of Environmental Health Sciences
Zooming In on Meiosis
Posted on by Dr. Francis Collins
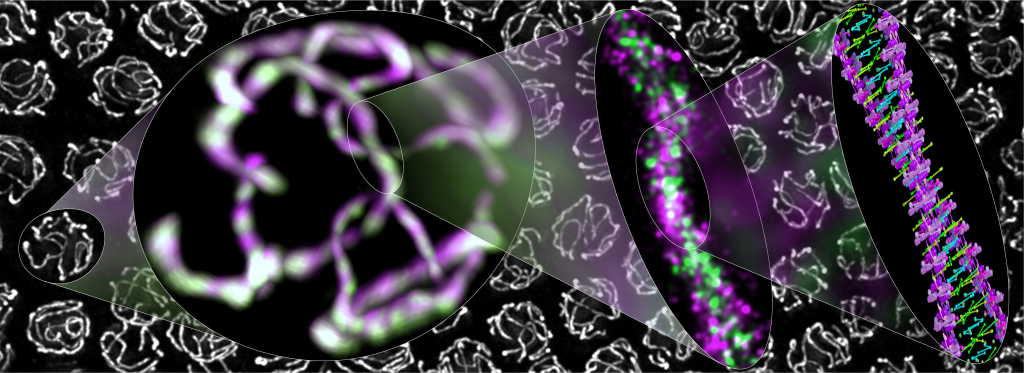
Credit: Simone Köhler, Michal Wojcik, Ke Xu, and Abby Dernburg, University of California, Berkeley
Meiosis—the formation of egg and sperm cells—is a highly choreographed process that creates genetic diversity in all plants and animals, including humans, to make each of us unique. This kaleidoscopic image shows cells from a worm exchanging DNA during meiosis.
You can see a protein-based polymer tether (green) from what’s called the synaptonemal complex. The complex holds together partner chromosomes (magenta) to facilitate DNA exchange in nuclei (white). Moving from left to right are views of the molecular assembly that progressively zoom in on the DNA, revealing in exquisite detail (far right) the two paired partner chromosomes perfectly aligned. This is not just the familiar DNA double helix. This is a double helix made up of two double helices!
Cryo-EM Images Capture Key Enzyme Tied to Cancer, Aging
Posted on by Dr. Francis Collins
Each time your cells divide, telomeres—complexes of specialized DNA sequences, RNA, and protein that protect the tips of your chromosomes—shorten just a bit. And, as the video shows, that shortening renders the genomic information on your chromosomes more vulnerable to changes that can drive cancer and other diseases of aging.
Consequently, over the last few decades, much research has focused on efforts to understand telomerase, a naturally occurring enzyme that helps to replace the bits of telomere lost during cell division. But there’s been a major hitch: until recently, scientists hadn’t been able to determine telomerase’s molecular structure in detail—a key step in figuring out exactly how the enzyme works. Now, thanks to better purification methods and an exciting technology called cryo-electron microscopy (cryo-EM), NIH-funded researchers and their colleagues have risen to the challenge to produce the most detailed view yet of human telomerase in its active form [1].
This structural biology advance is a critical step toward learning more about the role of telomerase in cancers, as well as genetic conditions linked to telomerase deficiencies. It’s also an important milestone in the quest for drugs targeting telomerase in different ways, perhaps to slow the growth of cancerous cells or to boost the proliferative capacity of life-giving adult stem cells.
One reason telomerase has been so difficult to study in humans is that the enzyme isn’t produced at detectable levels in the vast majority of our cells. To get around this problem, the team led by Eva Nogales and Kathleen Collins at the University of California, Berkeley, first coaxed human cells in the lab to produce larger quantities of active telomerase. They then used fluorescent microscopy, along with extensive knowledge of the enzyme’s biochemistry, to develop a multi-step purification process that yielded relatively homogenous samples of active telomerase.
The new study is also yet another remarkable example of how cryo-EM microscopy has opened up new realms of scientific possibility. That’s because, in comparison to other methods, cryo-EM enables researchers to solve complex macromolecular structures even when only tiny amounts of material are available. It can also produce detailed images of molecules, like telomerase, that are extremely flexible and hard to keep still while taking a picture of their structure.
As described in Nature, the researchers used cryo-EM to capture the structure of human telomerase in unprecedented detail. Their images reveal two lobes, held together by a flexible RNA tether. One of those lobes contains the highly specialized core enzyme. It uses an internal RNA template as a guide to make the repetitive, telomeric DNA that’s added at the tips of chromosomes. The second lobe, consisting of a complex of RNA and RNA-binding proteins, plays important roles in keeping the complex stable and properly in place.
This new, more-detailed view helps to explain how mutations in particular genes may lead to telomerase-related health conditions, including bone marrow failure, as well as certain forms of anemia and pulmonary fibrosis. For example, it reveals that a genetic defect known to cause bone marrow failure affects an essential protein in a spot that’s especially critical for telomerase’s proper conformation and function.
This advance will also be a big help for designing therapies that encourage telomerase activity. For example, it could help to boost the success of bone marrow transplants by rejuvenating adult stem cells. It might also be possible to reinforce the immune systems of people with HIV infections. While telomerase-targeted treatments surely won’t stop people from growing old, new insights into this important enzyme will help to understand aging better, including why some people appear to age faster than others.
As remarkable as these new images are, the researchers aren’t yet satisfied. They’ll continue to refine them down to the minutest structural details. They say they’d also like to use cryo-EM to understand better how the complex attaches to chromosomes to extend telomeres. Each new advance in the level of atomic detail will not only make for amazing new videos, it will help to advance understanding of human biology in health, aging, and disease.
References:
[1] Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K. Nature. 2018 April 25. [Epub ahead of publication]
Links:
High Resolution Electron Microscopy (National Cancer Institute/NIH)
Nogales Lab (University of California, Berkeley)
Collins Lab (University of California, Berkeley)
NIH Support: National Institute of General Medical Sciences
Shattering News: How Chromothripsis Cured a Rare Disease
Posted on by Dr. Francis Collins
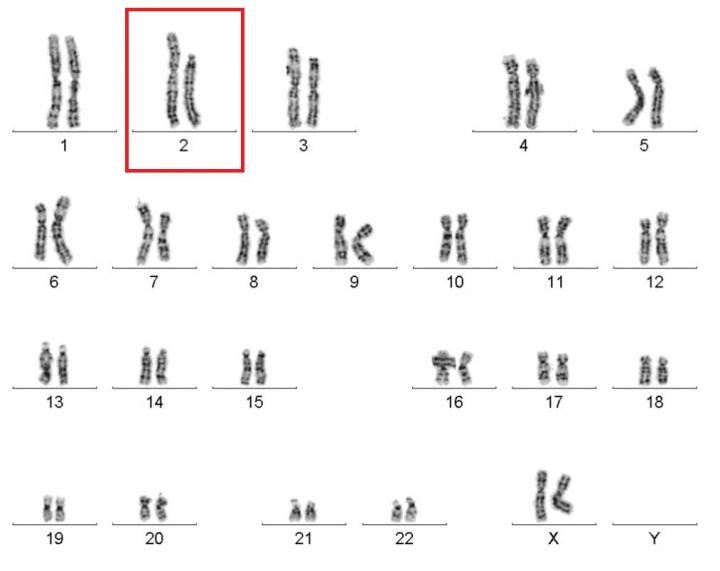
Caption: Karyotype of a woman spontaneously cured of WHIM syndrome. These chromosome pairings, which are from her white blood cells, show a normal chromosome 2 on the left, and a truncated chromosome 2 on the right.
Source: National Institute of Allergy and Infectious Diseases , NIH
The world of biomedical research is filled with surprises. Here’s a remarkable one published recently in the journal Cell [1]. A child born in the 1950s with a rare genetic immunodeficiency syndrome amazingly cured herself years later when part of one of her chromosomes spontaneously shattered into 18 pieces during replication of a blood stem cell. The damaged chromosome randomly reassembled, sort of like piecing together a broken vase, but it was still missing a shard of 164 genes—including the very gene that caused her condition.
Researchers say the chromosomal shattering probably took place in a cell in the bone marrow. The stem cell, now without the disease-causing gene, repopulated her immune system with healthy bone marrow-derived immune cells, resulting in cure of the syndrome.