GFP
The Amazing Brain: Tracking Molecular Events with Calling Cards
Posted on by Dr. Francis Collins
In days mostly gone by, it was fashionable in some circles for people to hand out calling cards to mark their arrival at special social events. This genteel human tradition is now being adapted to the lab to allow certain benign viruses to issue their own high-tech calling cards and mark their arrival at precise locations in the genome. These special locations show where there’s activity involving transcription factors, specialized proteins that switch genes on and off and help determine cell fate.
The idea is that myriad, well-placed calling cards can track brain development over time in mice and detect changes in transcription factor activity associated with certain neuropsychiatric disorders. This colorful image, which won first place in this year’s Show Us Your BRAINs! Photo and Video contest, provides a striking display of these calling cards in action in living brain tissue.
The image comes from Allen Yen, a PhD candidate in the lab of Joseph Dougherty, collaborating with the nearby lab of Rob Mitra. Both labs are located in the Washington University School of Medicine, St. Louis.
Yen and colleagues zoomed in on this section of mouse brain tissue under a microscope to capture dozens of detailed images that they then stitched together to create this high-resolution overview. The image shows neural cells (red) and cell nuclei (blue). But focus in on the neural cells (green) concentrated in the brain’s outer cortex (top) and hippocampus (two lobes in the upper center). They’ve been labelled with calling cards that were dropped off by adeno-associated virus [1].
Once dropped off, a calling card doesn’t bear a pretentious name or title. Rather, the calling card, is a small mobile snippet of DNA called a transposon. It gets dropped off with the other essential component of the technology: a specialized enzyme called a transposase, which the researchers fuse to one of many specific transcription factors of interest.
Each time one of these transcription factors of interest binds DNA to help turn a gene on or off, the attached transposase “grabs” a transposon calling card and inserts it into the genome. As a result, it leaves behind a permanent record of the interaction.
What’s also nice is the calling cards are programmed to give away their general locations. That’s because they encode a fluorescent marker (in this image, it’s a green fluorescent protein). In fact, Yen and colleagues could look under a microscope and tell from all the green that their calling card technology was in place and working as intended.
The final step, though, was to find out precisely where in the genome those calling cards had been left. For this, the researchers used next-generation sequencing to produce a cumulative history and map of each and every calling card dropped off in the genome.
These comprehensive maps allow them to identify important DNA-protein binding events well after the fact. This innovative technology also enables scientists to attribute past molecular interactions with observable developmental outcomes in a way that isn’t otherwise possible.
While the Mitra and Dougherty labs continue to improve upon this technology, it’s already readily adaptable to answer many important questions about the brain and brain disorders. In fact, Yen is now applying the technology to study neurodevelopment in mouse models of neuropsychiatric disorders, specifically autism spectrum disorder (ASD) [2]. This calling card technology also is available for any lab to deploy for studying a transcription factor of interest.
This research is supported by the Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative. One of the major goals of BRAIN Initiative is to accelerate the development and application of innovative technologies to gain new understanding of the brain. This award-winning image is certainly a prime example of striving to meet this goal. I’ll look forward to what these calling cards will tell us in the future about ASD and other important neurodevelopmental conditions affecting the brain.
References:
[1] A viral toolkit for recording transcription factor-DNA interactions in live mouse tissues. Cammack AJ, Moudgil A, Chen J, Vasek MJ, Shabsovich M, McCullough K, Yen A, Lagunas T, Maloney SE, He J, Chen X, Hooda M, Wilkinson MN, Miller TM, Mitra RD, Dougherty JD. Proc Natl Acad Sci U S A. 2020 May 5;117(18):10003-10014.
[2] A MYT1L Syndrome mouse model recapitulates patient phenotypes and reveals altered brain development due to disrupted neuronal maturation. Jiayang Chen, Mary E. Lambo, Xia Ge, Joshua T. Dearborn, Yating Liu, Katherine B. McCullough, Raylynn G. Swift, Dora R. Tabachnick, Lucy Tian, Kevin Noguchi, Joel R. Garbow, John N. Constantino. bioRxiv. May 27, 2021.
Links:
Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative (NIH)
Autism Spectrum Disorder (National Institute of Mental Health/NIH)
Dougherty Lab (Washington University School of Medicine, St. Louis)
Mitra Lab (Washington University School of Medicine)
Show Us Your BRAINs! Photo and Video Contest (BRAIN Initiative/NIH)
NIH Support: National Institute of Neurological Disorders and Stroke; National Institute of Mental Health; National Center for Advancing Translational Sciences; National Human Genome Research Institute; National Institute of General Medical Sciences
The Amazing Brain: Toward a Wiring Diagram of Connectivity
Posted on by Dr. Francis Collins
It’s summertime and, thanks to the gift of COVID-19 vaccines, many folks are getting the chance to take a break. So, I think it’s also time that my blog readers finally get a break from what’s been nearly 18 months of non-stop coverage of COVID-19 research. And I can’t think of a more enjoyable way to do that than by taking a look at just a few of the many spectacular images and insights that researchers have derived about the amazing brain.
The Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative, which is an NIH-led project aimed at revolutionizing our understanding of the human brain, happens to have generated some of the coolest—and most informative—imagery now available in neuroscience. So, throughout the month of August, I’ll share some of the entries from the initiative’s latest Show Us Your BRAINs! Photo and Video Contest.
With nearly 100 billion neurons and 100 trillion connections, the human brain remains one of the greatest mysteries in science. Among the many ways in which neuroscientists are using imaging to solve these mysteries is by developing more detailed maps of connectivity within the brain.
For example, the image featured above from the contest shows a dense weave of neurons in the anterior cingulate cortex, which is the part of the brain involved in learning, memory, and some motor control. In this fluorescence micrograph of tissue from a mouse, each neuron has been labeled with green fluorescent protein, enabling you to see how it connects to other neurons through arm-like projections called axons and dendrites.
The various connections, or circuits, within the brain process and relay distinct types of sensory information. In fact, a single neuron can form a thousand or more of these connections. Among the biggest challenges in biomedicine today is deciphering how these circuits work, and how they can misfire to cause potentially debilitating neurological conditions, including Alzheimer’s disease, Parkinson’s disease, autism, epilepsy, schizophrenia, depression, and traumatic brain injury.
This image was produced by Nicholas Foster and Lei Gao in the NIH-supported lab of Hong Wei Dong, University of California, Los Angeles. The Dong Lab is busy cataloging cell types and helping to assemble a wiring diagram of the connectivity in the mammalian brain—just one of the BRAIN Initiative’s many audacious goals. Stay tuned for more throughout the month of August!
Links:
Brain Research through Advancing Innovative Neurotechnologies® (BRAIN) Initiative (NIH)
Dong Lab (University of California, Los Angeles)
Show Us Your BRAINs! Photo and Video Contest (BRAIN Initiative/NIH)
NIH Support: National Institute of Mental Health
Finding Beauty in Cell Stress
Posted on by Dr. Francis Collins
Most stressful situations that we experience in daily life aren’t ones that we’d choose to repeat. But the cellular stress response captured in this video is certainly worth repeating a few times, so you can track what happens when two cancer cells get hit with stressors.
In this movie of two highly stressed osteosarcoma cells, you first see the appearance of many droplet-like structures (green). This is followed by a second set of droplets (magenta) and, finally, the fusion of both types of droplets.
These droplets are composed of fluorescently labeled stress-response proteins, either G3BP or UBQLN2 (Ubiquilin-2). Each protein is undergoing a fascinating process, called phase separation, in which a non-membrane bound compartment of the cytoplasm emerges and constrains the motion of proteins within it. Subsequently, the proteins fuse with like proteins to form larger droplets, in much the same way that raindrops merge on a car’s windshield.
Julia Riley, an undergraduate student in the NIH-supported lab of Heidi Hehnly and lab of Carlos Castañeda, Syracuse University, NY, shot this movie using the sophisticated tools of fluorescence microscopy. It’s the next installment in our series featuring winners of the 2019 Green Fluorescent Protein Image and Video Contest, sponsored by the American Society for Cell Biology. The contest honors the discovery of green fluorescent protein (GFP), which—together with a rainbow of other fluorescent proteins—has enabled researchers to visualize proteins and their dynamic activities inside cells for the last 25 years.
Riley and colleagues suspect that, in this case, phase separation is a protective measure that allows proteins to wall themselves off from the rest of the cell during stressful conditions. In this way, the proteins can create new functional units within the cell. The researchers are working to learn much more about what this interesting behavior entails as a basic organizing principle in the cell and how it works.
Even more intriguing is that similar stress-responding proteins are commonly altered in people with the devastating neurologic condition known as amyotrophic lateral sclerosis (ALS). ALS is a group of rare neurological diseases that involve the progressive deterioration of neurons responsible for voluntary movements such as chewing, walking, and talking. There’s been the suggestion that these phase separation droplets may seed the formation of the larger protein aggregates that accumulate in the motor neurons of people with this debilitating and fatal condition.
Castañeda and Hehnly, working with J. Paul Taylor at St. Jude Children’s Research Hospital, Memphis, earlier reported that Ubiquilin-2 forms stress-induced droplets in multiple cell types [1]. More recently, they showed that mutations in Ubiquilin-2 have been linked to ALS changes in the way that the protein undergoes phase separation in a test tube [2].
While the proteins in this award-winning video aren’t mutant forms, Riley is now working on the sequel, featuring versions of the Ubiquilin-2 protein that you’d find in some people with ALS. She hopes to capture how those mutations might produce a different movie and what that might mean for understanding ALS.
References:
[1] Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Dao TP, Kolaitis R-M, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, Castañeda CA. Molecular Cell. 2018 Mar 15;69(6):965-978.e6.
[2] ALS-Linked Mutations Affect UBQLN2 Oligomerization and Phase Separation in a Position- and Amino Acid-Dependent Manner. Dao TP, Martyniak B, Canning AJ, Lei Y, Colicino EG, Cosgrove MS, Hehnly H, Castañeda CA. Structure. 2019 Jun 4;27(6):937-951.e5.
Links:
Amyotrophic Lateral Sclerosis (ALS) (National Institute of Neurological Disorders and Stroke/NIH)
Castañeda Lab (Syracuse University, NY)
Hehnly Lab (Syracuse University)
Green Fluorescent Protein Image and Video Contest (American Society for Cell Biology, Bethesda, MD)
2008 Nobel Prize in Chemistry (Nobel Foundation, Stockholm, Sweden)
NIH Support: National Institute of General Medical Sciences
The Perfect Cytoskeletal Storm
Posted on by Dr. Francis Collins
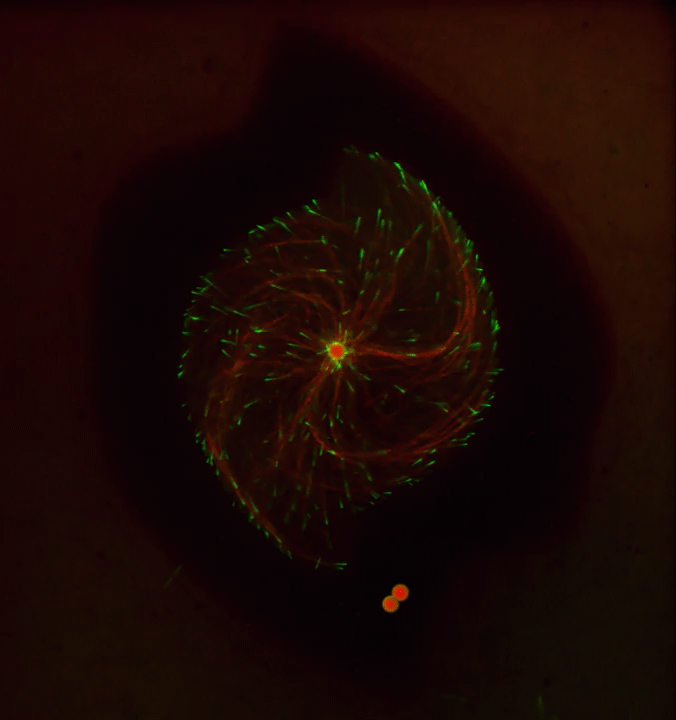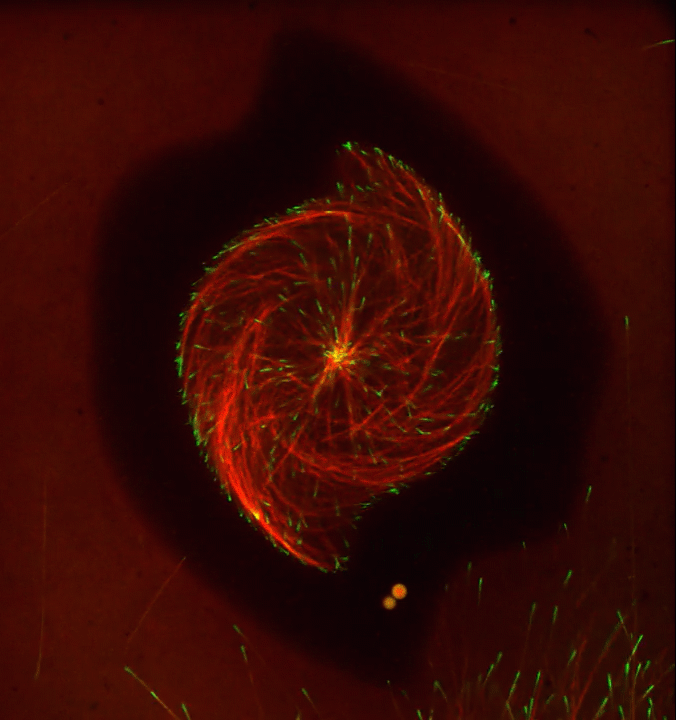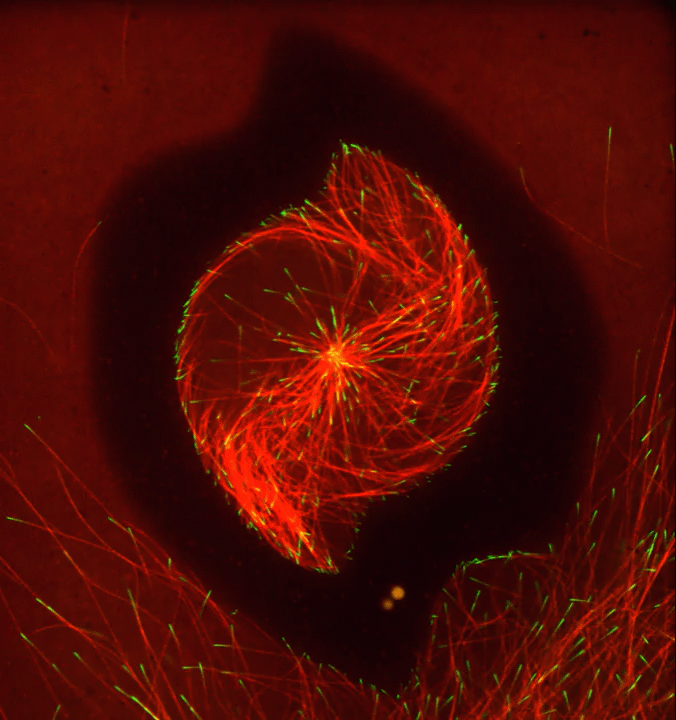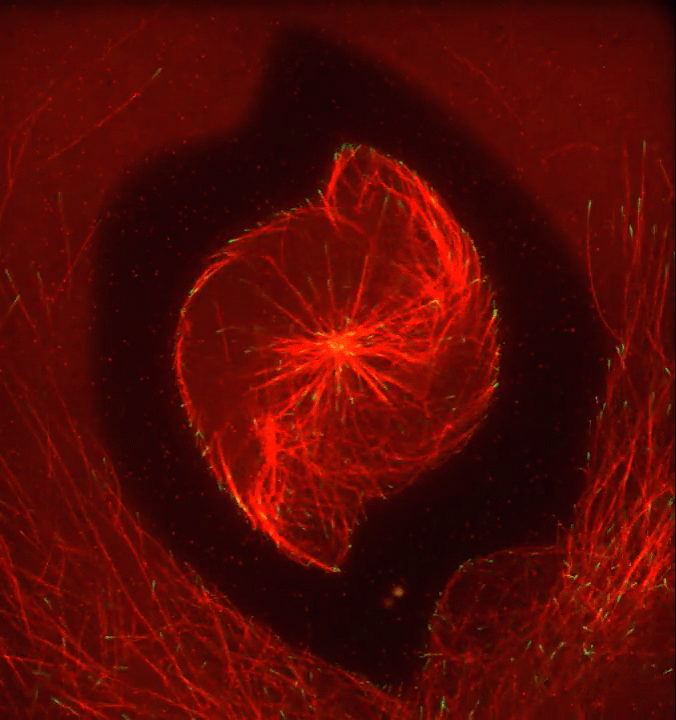
Ever thought about giving cell biology a whirl? If so, I suggest you sit down and take a look at this full-blown cytoskeletal “storm,” which provides a spectacular dynamic view of the choreography of life.
Before a cell divides, it undergoes a process called mitosis that copies its chromosomes and produces two identical nuclei. As part of this process, microtubules, which are structural proteins that help make up the cell’s cytoskeleton, reorganize the newly copied chromosomes into a dense, football-shaped spindle. The position of this mitotic spindle tells the cell where to divide, allowing each daughter cell to contain its own identical set of DNA.
To gain a more detailed view of microtubules in action, researchers designed an experimental system that utilizes an extract of cells from the African clawed frog (Xenopus laevis). As the video begins, a star-like array of microtubules (red) radiate outward in an apparent effort to prepare for cell division. In this configuration, the microtubules continually adjust their lengths with the help of the protein EB-1 (green) at their tips. As the microtubules grow and bump into the walls of a lab-generated, jelly-textured enclosure (dark outline), they buckle—and the whole array then whirls around the center.
Abdullah Bashar Sami, a Ph.D. student in the NIH-supported lab of Jesse “Jay” Gatlin, University of Wyoming, Laramie, shot this movie as a part his basic research to explore the still poorly understood physical forces generated by microtubules. The movie won first place in the 2019 Green Fluorescent Protein Image and Video Contest sponsored by the American Society for Cell Biology. The contest honors the 25th anniversary of the discovery of green fluorescent protein (GFP), which transformed cell biology and earned the 2008 Nobel Prize in Chemistry for three scientists who had been supported by NIH.
Like many movies, the setting was key to this video’s success. The video was shot inside a microfluidic chamber, designed in the Gatlin lab, to study the physics of microtubule assembly just before cells divide. The tiny chamber holds a liquid droplet filled with the cell extract.
When the liquid is exposed to an ultra-thin beam of light, it forms a jelly-textured wall, which traps the molecular contents inside [1]. Then, using time-lapse microscopy, the researchers watch the mechanical behavior of GFP-labeled microtubules [2] to see how they work to position the mitotic spindle. To do this, microtubules act like shapeshifters—scaling to adjust to differences in cell size and geometry.
The Gatlin lab is continuing to use their X. laevis system to ask fundamental questions about microtubule assembly. For many decades, both GFP and this amphibian model have provided cell biologists with important insights into the choreography of life, and, as this work shows, we can expect much more to come!
References:
[1] Microtubule growth rates are sensitive to global and local changes in microtubule plus-end density. Geisterfer ZM, Zhu D, Mitchison T, Oakey J, Gatlin JC. November 20, 2019.
[2] Tau-based fluorescent protein fusions to visualize microtubules. Mooney P, Sulerud T, Pelletier JF, Dilsaver MR, et al. Cytoskeleton (Hoboken). 2017 Jun;74(6):221-232.
Links:
Mitosis (National Human Genome Research Institute/NIH)
Gatlin Lab (University of Wyoming, Laramie)
Green Fluorescent Protein Image and Video Contest (American Society for Cell Biology, Bethesda, MD)
2008 Nobel Prize in Chemistry (Nobel Foundation, Stockholm, Sweden)
NIH Support: National Institute of General Medical Sciences
Seeing the Cytoskeleton in a Whole New Light
Posted on by Dr. Francis Collins

It’s been 25 years since researchers coaxed a bacterium to synthesize an unusual jellyfish protein that fluoresced bright green when irradiated with blue light. Within months, another group had also fused this small green fluorescent protein (GFP) to larger proteins to make their whereabouts inside the cell come to light—like never before.
To mark the anniversary of this Nobel Prize-winning work and show off the rainbow of color that is now being used to illuminate the inner workings of the cell, the American Society for Cell Biology (ASCB) recently held its Green Fluorescent Protein Image and Video Contest. Over the next few months, my blog will feature some of the most eye-catching entries—starting with this video that will remind those who grew up in the 1980s of those plasma balls that, when touched, light up with a simulated bolt of colorful lightning.
This video, which took third place in the ASCB contest, shows the cytoskeleton of a frequently studied human breast cancer cell line. The cytoskeleton is made from protein structures called microtubules, made visible by fluorescently tagging a protein called doublecortin (orange). Filaments of another protein called actin (purple) are seen here as the fine meshwork in the cell periphery.
The cytoskeleton plays an important role in giving cells shape and structure. But it also allows a cell to move and divide. Indeed, the motion in this video shows that the complex network of cytoskeletal components is constantly being organized and reorganized in ways that researchers are still working hard to understand.
Jeffrey van Haren, Erasmus University Medical Center, Rotterdam, the Netherlands, shot this video using the tools of fluorescence microscopy when he was a postdoctoral researcher in the NIH-funded lab of Torsten Wittman, University of California, San Francisco.
All good movies have unusual plot twists, and that’s truly the case here. Though the researchers are using a breast cancer cell line, their primary interest is in the doublecortin protein, which is normally found in association with microtubules in the developing brain. In fact, in people with mutations in the gene that encodes this protein, neurons fail to migrate properly during development. The resulting condition, called lissencephaly, leads to epilepsy, cognitive disability, and other neurological problems.
Cancer cells don’t usually express doublecortin. But, in some of their initial studies, the Wittman team thought it would be much easier to visualize and study doublecortin in the cancer cells. And so, the researchers tagged doublecortin with an orange fluorescent protein, engineered its expression in the breast cancer cells, and van Haren started taking pictures.
This movie and others helped lead to the intriguing discovery that doublecortin binds to microtubules in some places and not others [1]. It appears to do so based on the ability to recognize and bind to certain microtubule geometries. The researchers have since moved on to studies in cultured neurons.
This video is certainly a good example of the illuminating power of fluorescent proteins: enabling us to see cells and their cytoskeletons as incredibly dynamic, constantly moving entities. And, if you’d like to see much more where this came from, consider visiting van Haren’s Twitter gallery of microtubule videos here:
Reference:
[1] Doublecortin is excluded from growing microtubule ends and recognizes the GDP-microtubule lattice. Ettinger A, van Haren J, Ribeiro SA, Wittmann T. Curr Biol. 2016 Jun 20;26(12):1549-1555.
Links:
Lissencephaly Information Page (National Institute of Neurological Disorders and Stroke/NIH)
Wittman Lab (University of California, San Francisco)
Green Fluorescent Protein Image and Video Contest (American Society for Cell Biology, Bethesda, MD)
NIH Support: National Institute of General Medical Sciences
