zebrafish
Visit the New NIH Virtual Tour
Posted on by Lawrence Tabak, D.D.S., Ph.D.
Happy Fourth of July! Before everyone heads out to celebrate the holiday with their family and friends, I want to share this brief video with you. It’s an introduction to the brand-new NIH Virtual Tour that’s now available on our website. When time permits, I encourage everyone to take the full tour of our Bethesda, MD, main campus and explore this great institution of science, technological innovation, and, above all, hope.
Among the virtual tour’s many features is an interactive, aerial map of the 32 buildings on our Bethesda campus. By clicking on a highlighted building, you can explore an impressive multimedia gallery of photos, video clips, and other resources. The tour will allow you to learn more about NIH and the ways in which we help people live longer and healthier lives.
You also can learn more about NIH’s 27 Institutes and Centers, including the NIH Clinical Center and 20 other in-depth tour stops—from research labs to patient rooms—and hear directly from some of our impressive researchers, leaders, and patients. For example, you can learn about chronic pain research from a lab in the NIH Clinical Center or see the largest zebrafish facility in the world, housed in Building 6.
What I like most about the virtual tour is that it captures what makes NIH so special—the many amazing people who collaborate every day to discover ways to solve seemingly intractable research problems. I admire their commitment to follow the science wherever it may lead.
In fact, from its humble beginnings in a one-room laboratory in 1887, NIH has become the world’s largest funder of medical research, whether that’s mobilizing to combat a deadly pandemic or strategizing to help people with a rare disorder find answers.
Not only does NIH conduct groundbreaking research in its own labs and clinics, it also supports much of the medical research conducted at universities and institutions in your states and local communities. Whether in Bethesda or beyond the Beltway, this national research effort will continue to yield the needed understanding to turn discovery into better health, helping more people to flourish and lead fully productive lives, now and in the generations to come.
That’s certainly something we can all celebrate this holiday, the 247th birthday of our great nation that I’m so honored to serve. Have a great, but safe, Fourth of July, and I’ll see you back here soon to share another blog post and another story of NIH-supported research progress.
Links:
Virtual Tour (NIH)
Visitor Information (NIH)
Study in Africa Yields New Diabetes Gene
Posted on by Dr. Francis Collins
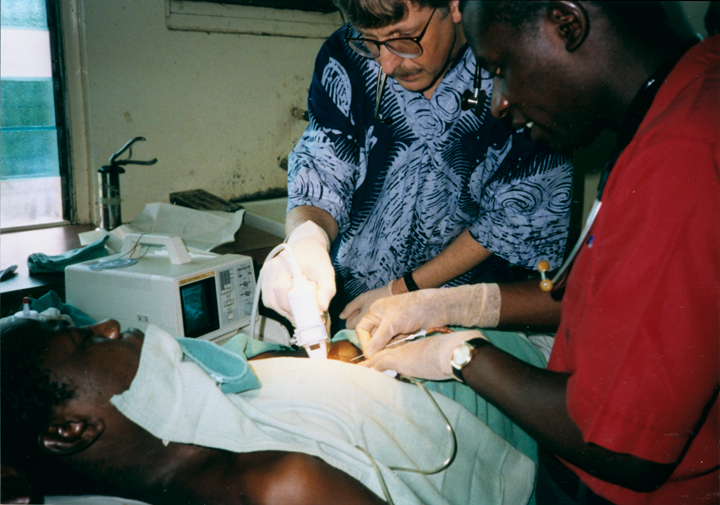
When I volunteered to serve as a physician at a hospital in rural Nigeria more than 25 years ago, I expected to treat a lot of folks with infectious diseases, such as malaria and tuberculosis. And that certainly happened. What I didn’t expect was how many people needed care for type 2 diabetes (T2D) and the health problems it causes. Surprisingly, these individuals were generally not overweight, and the course of their illness seemed different than in the West.
The experience inspired me to join with other colleagues at Howard University, Washington, DC, to help found the Africa America Diabetes Mellitus (AADM) study. It aims to uncover genomic risk factors for T2D in Africa and, using that information, improve understanding of the condition around the world.
So, I’m pleased to report that, using genomic data from more than 5,000 volunteers, our AADM team recently discovered a new gene, called ZRANB3, that harbors a variant associated with T2D in sub-Saharan Africa [1]. Using sophisticated laboratory models, the team showed that a malfunctioning ZRANB3 gene impairs insulin production to control glucose levels in the bloodstream.
Since my first trip to Nigeria, the number of people with T2D has continued to rise. It’s now estimated that about 8 to 10 percent of Nigerians have some form of diabetes [2]. In Africa, diabetes affects more than 7 percent of the population, more than twice the incidence in 1980 [3].
The causes of T2D involve a complex interplay of genetic, environmental, and lifestyle factors. I was particularly interested in finding out whether the genetic factors for T2D might be different in sub-Saharan Africa than in the West. But at the time, there was a dearth of genomic information about T2D in Africa, the cradle of humanity. To understand complex diseases like T2D fully, we need all peoples and continents represented in the research.
To begin to fill this research gap, the AADM team got underway and hasn’t looked back. In the latest study, led by Charles Rotimi at NIH’s National Human Genome Research Institute, in partnership with multiple African diabetes experts, the AADM team enlisted 5,231 volunteers from Nigeria, Ghana, and Kenya. About half of the study’s participants had T2D and half did not.
As reported in Nature Communications, their genome-wide search for T2D gene variants turned up three interesting finds. Two were in genes previously linked to T2D risk in other human populations. The third involved a gene that codes for ZRANB3, an enzyme associated with DNA replication and repair that had never been reported in association with T2D.
To understand how ZRANB3 might influence a person’s risk for developing T2D, the researchers turned to zebrafish (Danio rerio), an excellent vertebrate model for its rapid development. The researchers found that the ZRANB3 gene is active in insulin-producing beta cells of the pancreas. That was important to know because people with T2D frequently have reduced numbers of beta cells, which compromises their ability to produce enough insulin.
The team next used CRISPR/Cas9 gene-editing tools either to “knock out” or reduce the expression of ZRANB3 in young zebrafish. In both cases, it led to increased loss of beta cells.
Additional study in the beta cells of mice provided more details. While normal beta cells released insulin in response to high levels of glucose, those with suppressed ZRANB3 activity couldn’t. Together, the findings show that ZRANB3 is important for beta cells to survive and function normally. It stands to reason, then, that people with a lower functioning variant of ZRANB3 would be more susceptible to T2D.
In many cases, T2D can be managed with some combination of diet, exercise, and oral medications. But some people require insulin to manage the disease. The new findings suggest, particularly for people of African ancestry, that the variant of the ZRANB3 gene that one inherits might help to explain those differences. People carrying particular variants of this gene also may benefit from beginning insulin treatment earlier, before their beta cells have been depleted.
So why wasn’t ZRANB3 discovered in the many studies on T2D carried out in the United States, Europe, and Asia? It turns out that the variant that predisposes Africans to this disease is extremely rare in these other populations. Only by studying Africans could this insight be uncovered.
More than 20 years ago, I helped to start the AADM project to learn more about the genetic factors driving T2D in sub-Saharan Africa. Other dedicated AADM leaders have continued to build the research project, taking advantage of new technologies as they came along. It’s profoundly gratifying that this project has uncovered such an impressive new lead, revealing important aspects of human biology that otherwise would have been missed. The AADM team continues to enroll volunteers, and the coming years should bring even more discoveries about the genetic factors that contribute to T2D.
References:
[1] ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. Adeyemo AA, Zaghloul NA, Chen G, Doumatey AP, Leitch CC, Hostelley TL, Nesmith JE, Zhou J, Bentley AR, Shriner D, Fasanmade O, Okafor G, Eghan B Jr, Agyenim-Boateng K, Chandrasekharappa S, Adeleye J, Balogun W, Owusu S, Amoah A, Acheampong J, Johnson T, Oli J, Adebamowo C; South Africa Zulu Type 2 Diabetes Case-Control Study, Collins F, Dunston G, Rotimi CN. Nat Commun. 2019 Jul 19;10(1):3195.
[2] Diabetes mellitus in Nigeria: The past, present and future. Ogbera AO, Ekpebegh C. World J Diabetes. 2014 Dec 15;5(6):905-911.
[3] Global report on diabetes. Geneva: World Health Organization, 2016. World Health Organization.
Links:
Diabetes (National Institute of Diabetes ad Digestive and Kidney Diseases/NIH)
Diabetes and African Americans (Department of Health and Human Services)
Why Use Zebrafish to Study Human Diseases (Intramural Research Program/NIH)
Charles Rotimi (National Human Genome Research Institute/NIH)
NIH Support: National Human Genome Research Institute; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute on Minority Health and Health Disparities
Biomedical Research Highlighted in Science’s 2018 Breakthroughs
Posted on by Dr. Francis Collins

A Happy New Year to one and all! While many of us were busy wrapping presents, the journal Science announced its much-anticipated scientific breakthroughs of 2018. In case you missed the announcement [1], it was another banner year for the biomedical sciences.
The 2018 Breakthrough of the Year went to biomedical science and its ability to track the development of life—one cell at a time—in a variety of model organisms. This newfound ability opens opportunities to understand the biological basis of life more systematically than ever before. Among Science’s “runner-up” breakthroughs, more than half had strong ties to the biomedical sciences and NIH-supported research.
Sound intriguing? Let’s take a closer look at some of the amazing science conducted in 2018, starting with Science’s Breakthrough of the Year.
Development Cell by Cell: For millennia, biologists have wondered how a single cell develops into a complete multicellular organism, such as a frog or a mouse. But solving that mystery was almost impossible without the needed tools to study development systematically, one cell at a time. That’s finally started to change within the last decade. I’ve highlighted the emergence of some of these powerful tools on my blog and the interesting ways that they were being applied to study development.
Over the past few years, all of this technological progress has come to a head. Researchers, many of them NIH-supported, used sophisticated cell labeling techniques, nucleic acid sequencing, and computational strategies to isolate thousands of cells from developing organisms, sequence their genetic material, and determine their location within that developing organism.
In 2018 alone, groundbreaking single-cell analysis papers were published that sequentially tracked the 20-plus cell types that arise from a fertilized zebrafish egg, the early formation of organs in a frog, and even the creation of a new limb in the Axolotl salamander. This is just the start of amazing discoveries that will help to inform us of the steps, or sometimes missteps, within human development—and suggest the best ways to prevent the missteps. In fact, efforts are now underway to gain this detailed information in people, cell by cell, including the international Human Cell Atlas and the NIH-supported Human BioMolecular Atlas Program.
An RNA Drug Enters the Clinic: Twenty years ago, researchers Andrew Fire and Craig Mello showed that certain small, noncoding RNA molecules can selectively block genes in our cells from turning “on” through a process called RNA interference (RNAi). This work, for the which these NIH grantees received the 2006 Nobel Prize in Physiology or Medicine, soon sparked a wave of commercial interest in various noncoding RNA molecules for their potential to silence the expression of a disease-causing gene.
After much hard work, the first gene-silencing RNA drug finally came to market in 2018. It’s called Onpattro™ (patisiran), and the drug uses RNAi to treat the peripheral nerve disease that can afflict adults with a rare disease called hereditary transthyretin-mediated amyloidosis. This hard-won success may spark further development of this novel class of biopharmaceuticals to treat a variety of conditions, from cancer to cardiovascular disorders, with potentially greater precision.
Rapid Chemical Structure Determination: Last October, two research teams released papers almost simultaneously that described an incredibly fast new imaging technique to determine the structure of smaller organic chemical compounds, or “small molecules“ at atomic resolution. Small molecules are essential components of molecular biology, pharmacology, and drug development. In fact, most of our current medicines are small molecules.
News of these papers had many researchers buzzing, and I highlighted one of them on my blog. It described a technique called microcrystal electron diffraction, or MicroED. It enabled these NIH-supported researchers to take a powder form of small molecules (progesterone was one example) and generate high-resolution data on their chemical structures in less than a half-hour! The ease and speed of MicroED could revolutionize not only how researchers study various disease processes, but aid in pinpointing which of the vast number of small molecules can become successful therapeutics.
How Cells Marshal Their Contents: About a decade ago, researchers discovered that many proteins in our cells, especially when stressed, condense into circumscribed aqueous droplets. This so-called phase separation allows proteins to gather in higher concentrations and promote reactions with other proteins. The NIH soon began supporting several research teams in their groundbreaking efforts to explore the effects of phase separation on cell biology.
Over the past few years, work on phase separation has taken off. The research suggests that this phenomenon is critical in compartmentalizing chemical reactions within the cell without the need of partitioning membranes. In 2018 alone, several major papers were published, and the progress already has some suggesting that phase separation is not only a basic organizing principle of the cell, it’s one of the major recent breakthroughs in biology.
Forensic Genealogy Comes of Age: Last April, police in Sacramento, CA announced that they had arrested a suspect in the decades-long hunt for the notorious Golden State Killer. As exciting as the news was, doubly interesting was how they caught the accused killer. The police had the Golden Gate Killer’s DNA, but they couldn’t determine his identity, that is, until they got a hit on a DNA profile uploaded by one of his relatives to a public genealogy database.
Though forensic genealogy falls a little outside of our mission, NIH has helped to advance the gathering of family histories and using DNA to study genealogy. In fact, my blog featured NIH-supported work that succeeded in crowdsourcing 600 years of human history.
The researchers, using the online profiles of 86 million genealogy hobbyists with their permission, assembled more than 5 million family trees. The largest totaled more than 13 million people! By merging each tree from the crowd-sourced and public data, they were able to go back about 11 generations—to the 15th century and the days of Christopher Columbus. Though they may not have caught an accused killer, these large datasets provided some novel insights into our family structures, genes, and longevity.
An Ancient Human Hybrid: Every year, researchers excavate thousands of bone fragments from the remote Denisova Cave in Siberia. One such find would later be called Denisova 11, or “Denny” for short.
Oh, what a fascinating genomic tale Denny’s sliver of bone had to tell. Denny was at least 13 years old and lived in Siberia roughly 90,000 years ago. A few years ago, an international research team found that DNA from the mitochondria in Denny’s cells came from a Neanderthal, an extinct human relative.
In 2018, Denny’s family tree got even more interesting. The team published new data showing that Denny was female and, more importantly, she was a first generation mix of a Neanderthal mother and a father who belonged to another extinct human relative called the Denisovans. The Denisovans, by the way, are the first human relatives characterized almost completely on the basis of genomics. They diverged from Neanderthals about 390,000 years ago. Until about 40,000 years ago, the two occupied the Eurasian continent—Neanderthals to the west, and Denisovans to the east.
Denny’s unique genealogy makes her the first direct descendant ever discovered of two different groups of early humans. While NIH didn’t directly support this research, the sequencing of the Neanderthal genome provided an essential resource.
As exciting as these breakthroughs are, they only scratch the surface of ongoing progress in biomedical research. Every field of science is generating compelling breakthroughs filled with hope and the promise to improve the lives of millions of Americans. So let’s get started with 2019 and finish out this decade with more truly amazing science!
Reference:
[1] “2018 Breakthrough of the Year,” Science, 21 December 2018.
NIH Support: These breakthroughs represent the culmination of years of research involving many investigators and the support of multiple NIH institutes.
3D Action Film Stars Cancer Cell as the Villain
Posted on by Dr. Francis Collins
For centuries, microscopes have brought to light the otherwise invisible world of the cell. But microscopes don’t typically visualize the dynamic world of the cell within a living system.
For various technical reasons, researchers have typically had to displace cells, fix them in position, mount them onto slides, and look through a microscope’s viewfinder to see the cells. It can be a little like trying to study life in the ocean by observing a fish cooped up in an 8-gallon tank.
Now, a team partially funded by NIH has developed a new hybrid imaging technology to produce amazing, live-action 3D movies of living cells in their more natural state. In this video, you’re looking at a human breast cancer cell (green) making its way through a blood vessel (purple) of a young zebrafish.
At first, the cancer cell rolls along rather freely. As the cell adheres more tightly to the blood vessel wall, that rolling motion slows to a crawl. Ultimately, the cancer cell finds a place to begin making its way across and through the blood vessel wall, where it can invade other tissues.
Watching Cancer Cells Play Ball
Posted on by Dr. Francis Collins
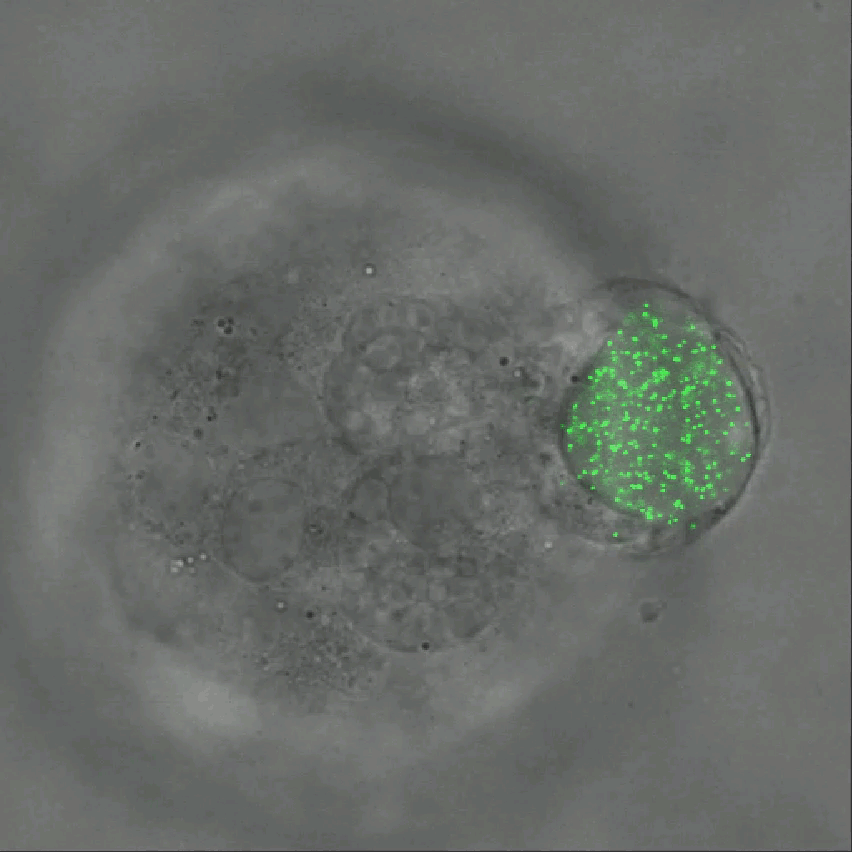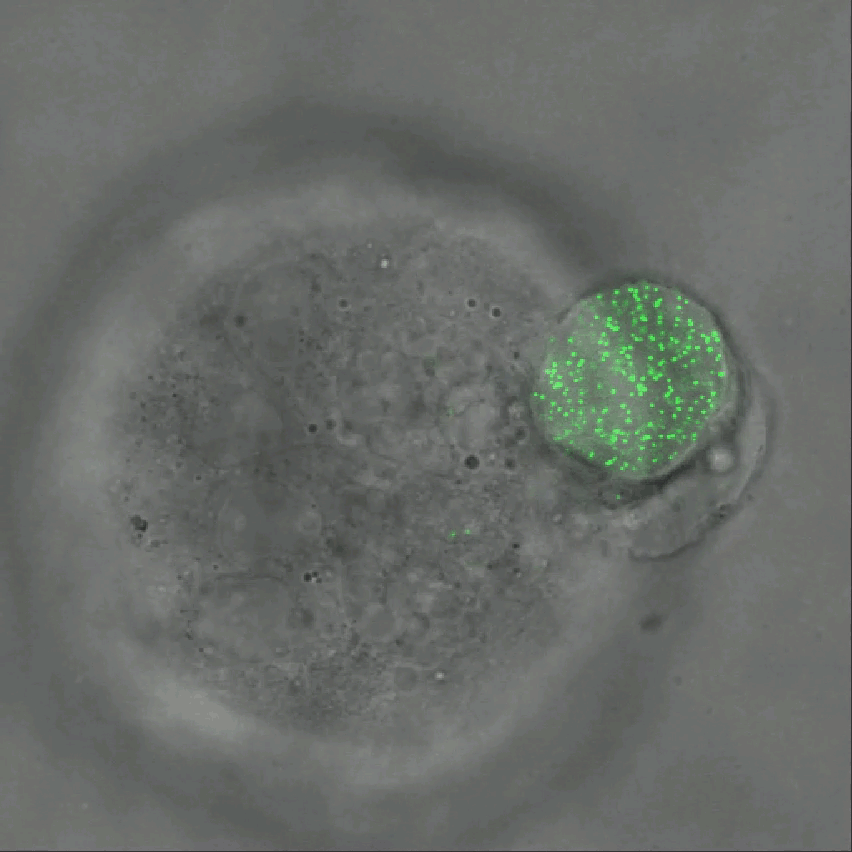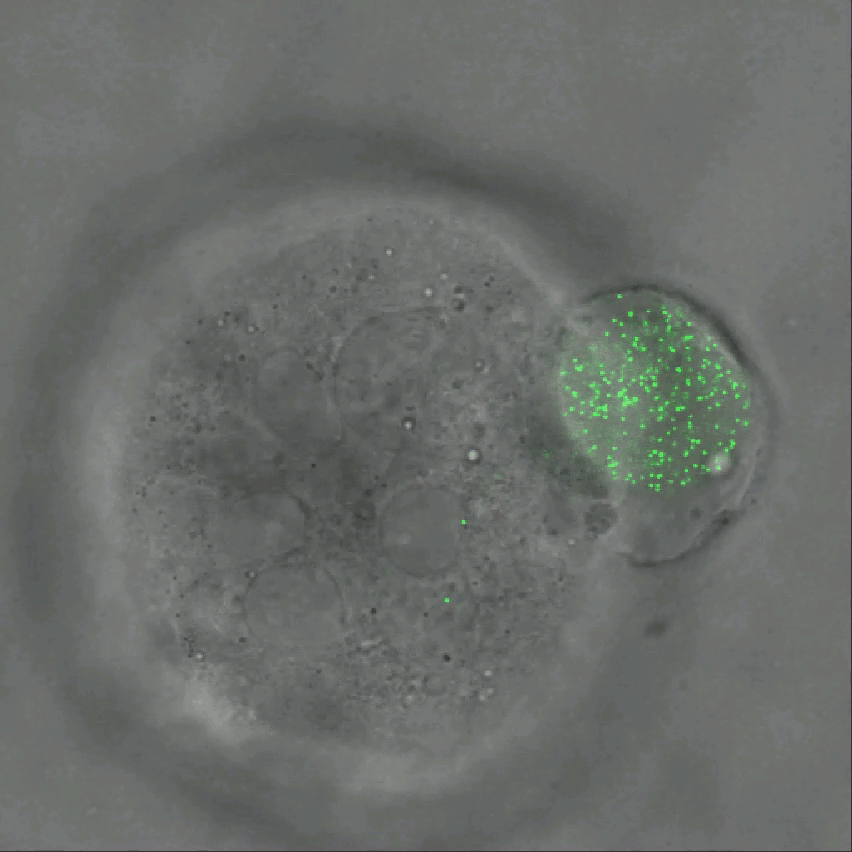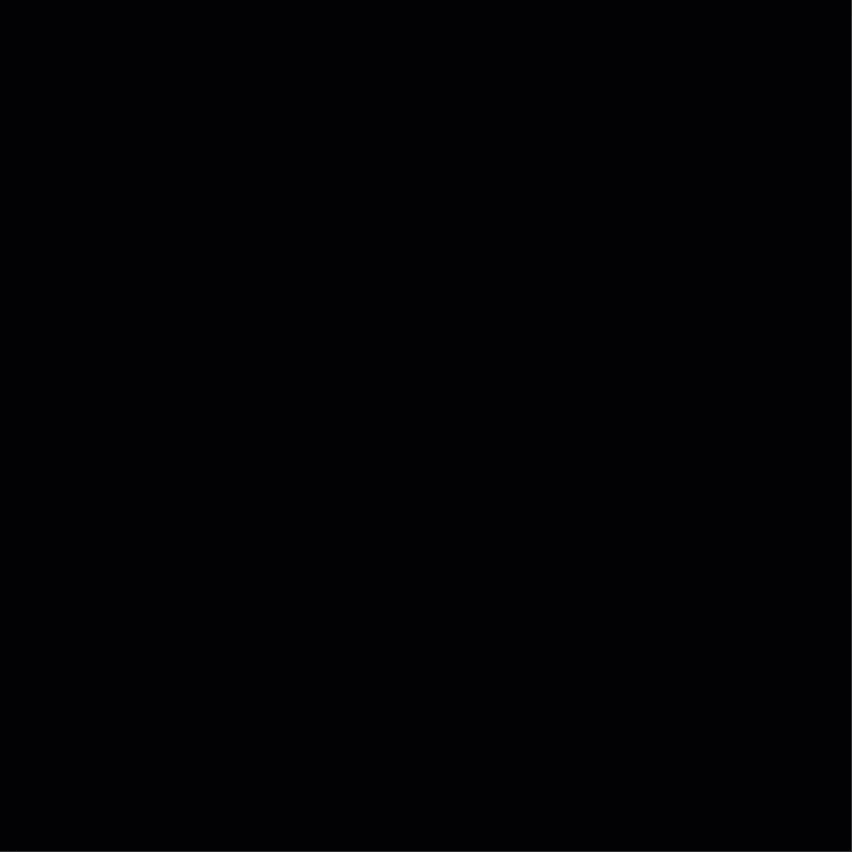
Credit: Ning Wang, University of Illinois at Urbana-Champaign
As tumor cells divide and grow, they push, pull, and squeeze one another. While scientists have suspected those mechanical stresses may play important roles in cancer, it’s been tough to figure out how. That’s in large part because there hadn’t been a good way to measure those forces within a tissue. Now, there is.
As described in Nature Communications, an NIH-funded research team has developed a technique for measuring those subtle mechanical forces in cancer and also during development [1]. Their ingenious approach is called the elastic round microgel (ERMG) method. It relies on round elastic microspheres—similar to miniature basketballs, only filled with fluorescent nanoparticles in place of air. In the time-lapse video above, you see growing and dividing melanoma cancer cells as they squeeze and spin one of those cell-sized “balls” over the course of 24 hours.
First Day in the Life of Nine Amazing Creatures
Posted on by Dr. Francis Collins
Credit: Tessa Montague, Harvard University, and Zuzka Vavrušová, University of California, San Francisco
Each summer for the last 125 years, students from around the country have traveled to the Marine Biological Laboratory (MBL), Woods Hole, MA, for an intensive course in embryology. While visiting this peaceful and scenic village on Cape Cod, they’re exposed to a dizzying array of organisms and state-of-the-art techniques to study their development.
Snapshots of Life: The Brain’s Microscopic Green Trash Bins
Posted on by Dr. Francis Collins

Credit: Marina Venero Galanternik, Daniel Castranova, Tuyet Nguyen, and Brant M. Weinstein, NICHD, NIH
There are trash bins in our homes, on our streets, and even as a popular icon on our desktop computers. And as this colorful image shows, trash bins of the cellular variety are also important in the brain.
This image—a winner in the Federation of American Societies for Experimental Biology’s 2017 BioArt competition—shows the brain of an adult zebrafish, a popular organism for studying how the brain works. It captures dense networks of blood vessels (red) lining the outer surface of the brain. Next to many of these vessels sit previously little-studied cells called fluorescent granular perithelial cells (yellowish green). Researchers now believe these cells, often shortened to FGPs, act much like trash receptacles that continuously take in and store waste products to keep the brain tidy and functioning well.
Finding Brain Circuits Tied to Alertness
Posted on by Dr. Francis Collins
Everybody knows that it’s important to stay alert behind the wheel or while out walking on the bike path. But our ability to react appropriately to sudden dangers is influenced by whether we feel momentarily tired, distracted, or anxious. How is it that the brain can transition through such different states of consciousness while performing the same routine task, even as its basic structure and internal wiring remain unchanged?
A team of NIH-funded researchers may have found an important clue in zebrafish, a popular organism for studying how the brain works. Using a powerful new method that allowed them to find and track brain circuits tied to alertness, the researchers discovered that this mental state doesn’t work like an on/off switch. Rather, alertness involves several distinct brain circuits working together to bring the brain to attention. As shown in the video above that was taken at cellular resolution, different types of neurons (green) secrete different kinds of chemical messengers across the zebrafish brain to affect the transition to alertness. The messengers shown are: serotonin (red), acetylcholine (blue-green), and dopamine and norepinephrine (yellow).
What’s also fascinating is the researchers found that many of the same neuronal cell types and brain circuits are essential to alertness in zebrafish and mice, despite the two organisms being only distantly related. That suggests these circuits are conserved through evolution as an early fight-or-flight survival behavior essential to life, and they are therefore likely to be important for controlling alertness in people too. If correct, it would tell us where to look in the brain to learn about alertness not only while doing routine stuff but possibly for understanding dysfunctional brain states, ranging from depression to post-traumatic stress disorder (PTSD).
Snapshots of Life: Coming Face to Face with Development
Posted on by Dr. Francis Collins
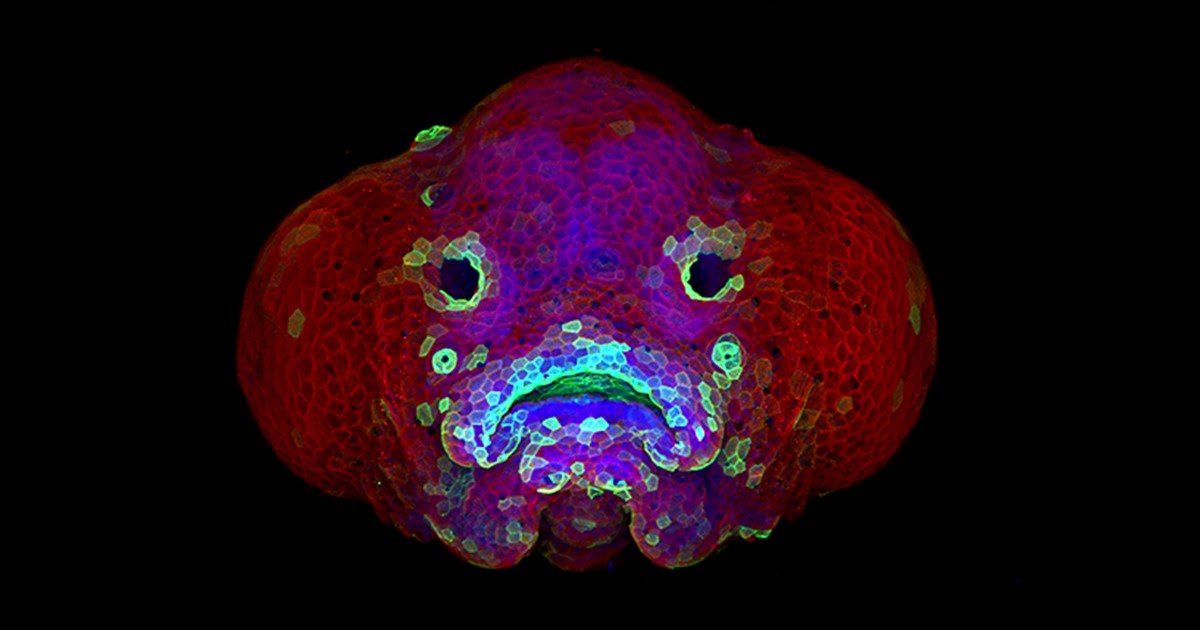
Credit: Oscar Ruiz and George Eisenhoffer, University of Texas MD Anderson Cancer Center, Houston
Zebrafish (Danio rerio) is a favorite model for studying development, in part because its transparent embryos make it possible to produce an ever-growing array of amazingly informative images. For one recent example, check out this Federation of American Societies for Experimental Biology’s 2016 BioArt winner, which shows the developing face of a 6-day-old zebrafish larva.
Yes, those downturned “lips” are indeed cells that will go on to become the fish’s mouth. But all is not quite what it appears: the two dark circles that look like eyes are actually developing nostrils. Both the nostrils and mouth express high levels of F-actin (green), a structural protein that helps orchestrate cell movement. Meanwhile, the two bulging areas on either side of the fish’s head, which are destined to become eyes and skin, express keratin (red).
Oscar Ruiz, who works in the lab of George Eisenhoffer at The University of Texas MD Anderson Cancer Center, Houston, used a confocal microscope to create this image. What was most innovative about his work was not the microscope itself, but how he prepared the sample for imaging. With traditional methods, researchers can only image the faces of zebrafish larvae from the side or the bottom. However, the Eisenhoffer lab has devised a new method of preparing fish larvae that makes it possible to image their faces head-on. This has enabled the team to visualize facial development at much higher resolution than was previously possible.
Next Page

