bioinformatics
National Library of Medicine Helps Lead the Way in AI Research
Posted on by Patricia Flatley Brennan, R.N., Ph.D., National Library of Medicine
Did you know that the NIH’s National Library of Medicine (NLM) has been serving science and society since 1836? From its humble beginning as a small collection of books in the library of the U.S. Army Surgeon General’s office, NLM has grown not only to become the world’s largest biomedical library, but a leader in biomedical informatics and computational health data science research.
Think of NLM as a door through which you pass to connect with health data, literature, medical and scientific information, expertise, and sophisticated mathematical models or images that describe a clinical problem. This intersection of information, people, and technology allows NLM to foster discovery. NLM does so by ensuring that scientists, clinicians, librarians, patients, and the public have access to biomedical information 24 hours a day, 7 days a week.
The NLM also supports two research efforts: the Division of Extramural Programs (EP) and Intramural Research Program (IRP). Both programs are accelerating advances in biomedical informatics, data science, computational biology, and computational health. One of EP’s notable investments is focused on advancing artificial intelligence (AI) methods and reimagining how health care is delivered with the power of AI.

With support from NLM, Corey Lester and his colleagues at the University of Michigan College of Pharmacy, Ann Arbor, MI, are using AI to assist in pill verification, a standard procedure in pharmacies across the land. They want to help pharmacists avoid dangerous and costly dispensing errors. To do so, Lester is using AI to develop a real-time computer vision model. It views pills inside of a medication bottle, accurately identifies them, and determines that they are the correct or incorrect contents.
The IRP develops and applies computational methods and approaches to a broad range of information problems in biology, biomedicine, and human health. The IRP also offers intramural training opportunities and supports other training aimed at pre-baccalaureate to postdoctoral students and professionals.
The NLM principal investigators use biological data to advance computer algorithms and connect relationships between any level of biological organization and health conditions. They also use computational health sciences to focus on clinical information processing and analyze clinical data, assess clinical outcomes, and set health data standards.

NLM investigator Sameer Antani is collaborating with researchers in other NIH institutes to explore how AI can help us understand oral cancer, echocardiography, and pediatric tuberculosis. His research also is examining how images can be mined for data to predict the causes and outcomes of conditions. Examples of Antani’s work can be found in mobile radiology vehicles, which allow professionals to take chest X-rays (right) and screen for HIV and tuberculosis using software containing algorithms developed in his lab.
For AI to have its full impact, more algorithms and approaches that harness the power of data are needed. That’s why NLM supports hundreds of other intramural and extramural scientists who are addressing challenging health and biomedical problems. The NLM-funded research is focused on how AI can help people stay healthy through early disease detection, disease management, and clinical and treatment decision-making—all leading to the ultimate goal of helping people live healthier and happier lives.
The NLM is proud to lead the way in the use of AI to accelerate discovery and transform health care. Want to learn more? Follow me on Twitter. Or, you can follow my blog, NLM Musings from the Mezzanine and receive periodic NLM research updates.
I would like to thank Valerie Florance, Acting Scientific Director of NLM IRP, and Richard Palmer, Acting Director of NLM Division of EP, for their assistance with this post.
Links:
National Library of Medicine (National Library of Medicine/NIH)
Video: Using Machine Intelligence to Prevent Medication Dispensing Errors (NLM Funding Spotlight)
Video: Sameer Antani and Artificial Intelligence (NLM)
NLM Division of Extramural Programs (NLM)
NLM Intramural Research Program (NLM)
NLM Intramural Training Opportunities (NLM)
Principal Investigators (NLM)
NLM Musings from the Mezzanine (NLM)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 20th in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
How COVID-19 Took Hold in North America and Europe
Posted on by Dr. Francis Collins
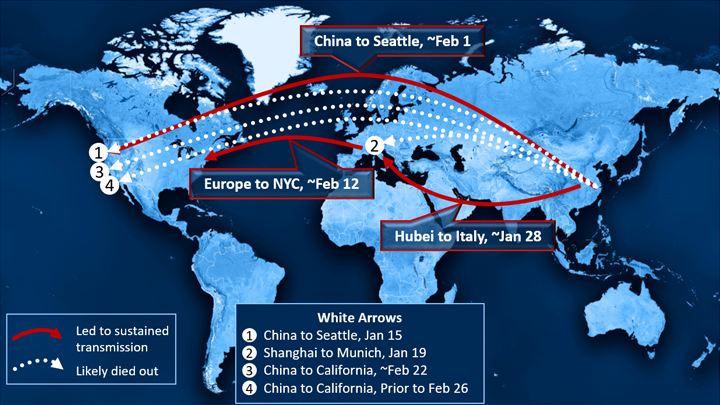
It was nearly 10 months ago on January 15 that a traveler returned home to the Seattle area after visiting family in Wuhan, China. A few days later, he started feeling poorly and became the first laboratory-confirmed case of coronavirus disease 2019 (COVID-19) in the United States. The rest is history.
However, new evidence published in the journal Science suggests that this first COVID-19 case on the West Coast didn’t snowball into the current epidemic. Instead, while public health officials in Washington state worked tirelessly and ultimately succeeded in containing its sustained transmission, the novel coronavirus slipped in via another individual about two weeks later, around the beginning of February.
COVID-19 is caused by the novel coronavirus SARS-CoV-2. Last winter, researchers sequenced the genetic material from the SARS-CoV-2 that was isolated from the returned Seattle traveler. While contact tracing didn’t identify any spread of this particular virus, dubbed WA1, questions arose when a genetically similar virus known as WA2 turned up in Washington state. Not long after, WA2-like viruses then appeared in California; British Columbia, Canada; and eventually 3,000 miles away in Connecticut. By mid-March, this WA2 cluster accounted for the vast majority—85 percent—of the cases in Washington state.
But was it possible that the WA2 cluster is a direct descendent of WA1? Did WA1 cause an unnoticed chain of transmission over several weeks, making the Seattle the epicenter of the outbreak in North America?
To answer those questions and others from around the globe, Michael Worobey, University of Arizona, Tucson, and his colleagues drew on multiple sources of information. These included data peretaining to viral genomes, airline passenger flow, and disease incidence in China’s Hubei Province and other places that likely would have influenced the probability that infected travelers were moving the virus around the globe. Based on all the evidence, the researchers simulated the outbreak more than 1,000 times on a computer over a two-month period, beginning on January 15 and assuming the epidemic started with WA1. And, not once did any of their simulated outbreaks match up to the actual genome data.
Those findings suggest to the researchers that the idea WA1 is responsible for all that came later is exceedingly unlikely. The evidence and simulations also appear to rule out the notion that the earliest cases in Washington state entered the United States by way of Canada. A deep dive into the data suggests a more likely scenario is that the outbreak was set off by one or more introductions of genetically similar viruses from China to the West Coast. Though we still don’t know exactly where, the Seattle area is the most likely site given the large number of WA2-like viruses sampled there.
Worobey’s team conducted a second analysis of the outbreak in Europe, and those simulations paint a similar picture to the one in the United States. The researchers conclude that the first known case of COVID-19 in Europe, arriving in Germany on January 20, led to a relatively small number of cases before being stamped out by aggressive testing and contact tracing efforts. That small, early outbreak probably didn’t spark the later one in Northern Italy, which eventually spread to the United States.
Their findings also show that the chain of transmission from China to Italy to New York City sparked outbreaks on the East Coast slightly later in February than those that spread from China directly to Washington state. It confirms that the Seattle outbreak was indeed the first, predating others on the East Coast and in California.
The findings in this report are yet another reminder of the value of integrating genome surveillance together with other sources of data when it comes to understanding, tracking, and containing the spread of COVID-19. They also show that swift and decisive public health measures to contain the virus worked when SARS-CoV-2 first entered the United States and Europe, and can now serve as models of containment.
Since the suffering and death from this pandemic continues in the United States, this historical reconstruction from early in 2020 is one more reminder that all of us have the opportunity and the responsibility to try to limit further spread. Wear your mask when you are outside the home; maintain physical distancing; wash your hands frequently; and don’t congregate indoors, where the risks are greatest. These lessons will enable us to better anticipate, prevent, and respond to additional outbreaks of COVID-19 or any other novel viruses that may arise in the future.
Reference:
[1] The emergence of SARS-CoV-2 in Europe and North America. Worobey M, Pekar J, Larsen BB, Nelson MI, Hill V, Joy JB, Rambaut A, Suchard MA, Wertheim JO, Lemey P. Science. 2020 Sep 10:eabc8169 [Epub ahead of print]
Links:
Coronavirus (COVID-19) (NIH)
Michael Worobey (University of Arizona, Tucson)
NIH Support: National Institute of Allergy and Infectious Diseases; Fogarty International Center; National Library of Medicine
Electricity-Conducting Bacteria May Inspire Next-Gen Medical Devices
Posted on by Dr. Francis Collins

Technological advances with potential for improving human health sometimes come from the most unexpected places. An intriguing example is an electricity-conducting biological nanowire that holds promise for powering miniaturized pacemakers and other implantable electronic devices.
The nanowires come from a bacterium called Geobacter sulfurreducens, shown in the electron micrograph above. This rod-shaped microbe (white) was discovered two decades ago in soil collected from an unlikely place: a ditch outside of Norman, Oklahoma. The bug can conduct electricity along its arm-like appendages, and, in the hydrocarbon-contaminated, oxygen-depleted soil in which it lives, such electrical inputs and outputs are essentially the equivalent of breathing.
Scientists fascinated with G. sulfurreducens thought that its electricity had to be flowing through well-studied microbial appendages called pili. But, as the atomic structure of these nanowires (multi-colors, foreground) now reveals, these nanowires aren’t pili at all! Instead, the bacteria have manufactured unique submicroscopic arm-like structures. These arms consist of long, repetitive chains of a unique protein, each surrounding a core of iron-containing molecules.
The surprising discovery, published in the journal Cell, was made by an NIH-funded team involving Edward Egelman, University of Virginia Health System, Charlottesville. Egelman’s lab has had a long interest in what’s called a type 4 pili. These strong, adhering appendages help certain infectious bacteria enter tissues and make people sick. In fact, they enable bugs like Neisseria meningitidis to cross the blood-brain barrier and cause potentially deadly bacterial meningitis. While other researchers had proposed that those same type 4 pili allowed G. sulfurreducens to conduct electricity, Egelman wasn’t so sure.
So, he took advantage of recent advances in cryo-electron microscopy, which involves flash-freezing molecules at extremely low temperatures before bombarding them with electrons to capture their images with a special camera. The cryo-EM images allowed his team to nail down the atomic structure of the nanowires, now called OmcS filaments.
Using those images and sophisticated bioinformatics, Egelman and team determined that OmcS proteins uniquely fit into the nanowires’ long repetitive chains, spacing their iron-bearing cores at regular intervals to transfer electrons and convey electricity. In fact, bacteria unable to produce OmcS proteins make filaments that conduct electricity 100 times less efficiently.
With these cryo-EM structures in hand, Egelman says his team will continue to explore their conductive properties. Such knowledge might someday be used to build biologically-inspired nanowires, measuring 1/100,000th the width of a human hair, to connect miniature electronic devices directly to living tissues. This is one more example of how nature’s ability to invent is pretty breathtaking—surely one wouldn’t have predicted the discovery of nanowires in a bacterium that lives in contaminated ditches.
Reference:
[1] Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Wang F, Gu Y, O’Brien JP, Yi SM, Yalcin SE, Srikanth V, Shen C, Vu D, Ing NL, Hochbaum AI, Egelman EH, Malvankar NS. Cell. 2019 Apr 4;177(2):361-369.
Links:
Electroactive microorganisms in bioelectrochemical systems. Logan BE, Rossi R, Ragab A, Saikaly PE. Nat Rev Microbiol. 2019 May;17(5):307-319.
High Resolution Electron Microscopy (National Cancer Institute/NIH)
Egelman Lab (University of Virginia, Charlottesville)
NIH Support: National Institute of General Medical Sciences; National Institute of Allergy and Infectious Diseases; Common Fund
Creative Minds: Looking for Common Threads in Rare Diseases
Posted on by Dr. Francis Collins

Valerie Arboleda
Credit: UCLA/Margaret Sison Photography
Four years ago, Valerie Arboleda accomplished something most young medical geneticists rarely do. She helped discover a rare congenital disease now known as KAT6A syndrome [1]. From the original 10 cases to the more than 100 diagnosed today, KAT6A kids share a single altered gene that causes neuro-developmental delays, most prominently in learning to walk and talk, plus a spectrum of possible abnormalities involving the head, face, heart, and immune system.
Now, Arboleda wants to accomplish something even more groundbreaking. With a 2017 NIH Director’s Early Independence Award, she will develop ways to mine Big Data—the voluminous amounts of DNA sequence and other biological information now stored in public databases—to unearth new clues into the biology of rare disorders like KAT6A syndrome. If successful, Arboleda’s work could bring greater precision to the diagnosis and potentially treatment of Mendelian disorders, as well as provide greater clarity into the specific challenges that might lie ahead for an affected child.
Snapshots of Life: Imperfect but Beautiful Intruder
Posted on by Dr. Francis Collins
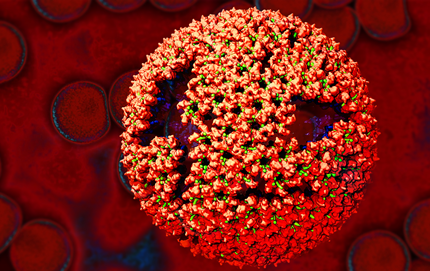
Credit: Boon Chong Goh, Beckman Institute, University of Illinois at Urbana-Champaign
The striking image you see above is an example of what can happen when scientists combine something old with something new. In this case, a researcher took the Rous sarcoma virus (RSV)—a virus that’s been studied for more than century because of its ability to cause cancer in chickens and the insights it provided on human oncogenes [1, 2]—and used modern computational tools to generate a model of its atomic structure.
Here you see an immature RSV particle that’s just budded from an infected chicken cell and entered the avian bloodstream. A lattice of proteins (red) held together by short peptides (green) cover the outer shell of the immature virus, shielding other proteins (blue) that make up an inner shell.