rheumatoid arthritis
Study Offers New Clues to Why Most People with Autoimmune Diseases Are Women
Posted on by Dr. Monica M. Bertagnolli
As many as 50 million Americans have one of more than 100 known autoimmune diseases, making it the third most prevalent disease category, surpassed only by cancer and heart disease.1,2 This category of disease has also long held a mystery: Why are most people with a chronic autoimmune condition—as many as four out of every five—women? This sex-biased trend includes autoimmune diseases such as rheumatoid arthritis, multiple sclerosis, scleroderma, lupus, Sjögren’s syndrome, and many others.
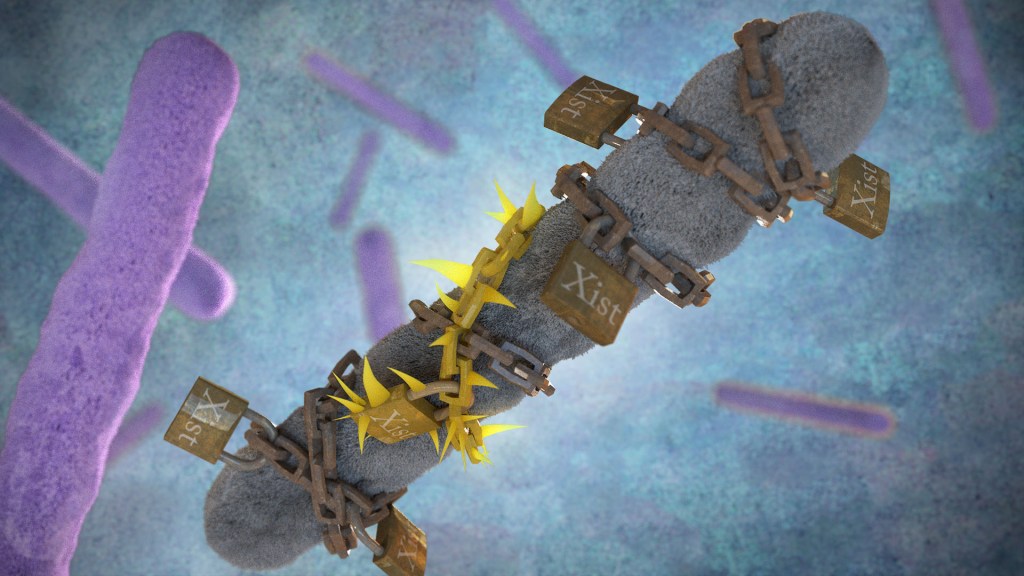
Now, exciting findings from a study supported in part by NIH provide a clue to why this may be the case, with potentially important implications for the early detection, treatment, and prevention of autoimmune diseases. The new evidence, reported in the journal Cell, suggests that more women develop autoimmune diseases than men due in part to the most fundamental difference between the biological sexes: that females have two X chromosomes, while males have an X and a Y. More specifically, it has to do with molecules called Xist (pronounced “exist”), which are encoded on the X chromosome and transcribed into long non-coding stretches of RNA, only when there are two X chromosomes.
Those long Xist molecules wind themselves around sections of just one of a female’s two X chromosomes, shutting down the extra X chromosome in a process known as X-chromosome inactivation. It’s an essential process to ensure those cells won’t produce too many proteins encoded on X chromosomes, which would be a deadly mistake. It’s also something that males, with a single X chromosome and much smaller Y chromosome carrying almost no working genes, don’t have to worry about.
The new findings come from a team at Stanford University School of Medicine, Stanford, CA, led by Howard Chang and Diana Dou. What they suggest is that while Xist molecules play an essential role in X-chromosome inactivation, they also have a more nefarious ability to encourage the formation of odd clumps of RNA, DNA, and proteins that can in turn trigger strong autoimmune responses.
In earlier research, the team identified about 80 different proteins that bind to Xist either directly or indirectly. After taking a close look at the list, the researchers realized that many of the proteins had been shown to play some role in autoimmune conditions. This raised an intriguing question: Could the reason women develop autoimmune diseases so much more often than men be explained by those Xist-containing clumps?
To test the idea, the researchers first decided to study it in male mice. They made two different strains of male mice produce Xist to see if it would increase their risk for autoimmunity in ways they could measure. And it did. The researchers found that once Xist was activated in male mice that were genetically prone to autoimmunity, they became more susceptible to developing a lupus-like condition. It didn’t happen in every individual, which suggests, not surprisingly, that the development of autoimmune disease requires additional triggers as well.
In addition, in a different mouse strain that was resistant to developing autoimmunity, the addition of Xist in males wasn’t enough to cause autoimmunity, the researchers found. That also makes sense in that, while women are much more prone to developing autoimmune disease, most people don’t. Xist complexes likely lead to autoimmunity only when certain genetic and other factors are met.
The researchers also examined blood samples from 100 people with autoimmune conditions and found they had antibodies to many of their own Xist complexes. Some of those antibodies also appeared specific to a certain autoimmune disorder, suggesting that they might be useful for tests that could detect autoimmunity or particular autoimmune conditions even before symptoms arise.
There are still many questions to explore in future research, including why men sometimes do get autoimmune conditions, and what other key triggers drive the development of autoimmunity. But this fundamentally important discovery points to potentially new ways to think about the causes for the autoimmune conditions that affect so many people in communities here and around the world.
References:
[1] The American Autoimmune Related Diseases Association. Autoimmune Facts.
[2] Dou DR, et al. Xist ribonucleoproteins promote female sex-biased autoimmunity. Cell. DOI: 10.1016/j.cell.2023.12.037. (2024).
NIH Support: National Institute of Arthritis and Musculoskeletal and Skin Diseases
Connecting the Dots: Oral Infection to Rheumatoid Arthritis
Posted on by Lawrence Tabak, D.D.S., Ph.D.
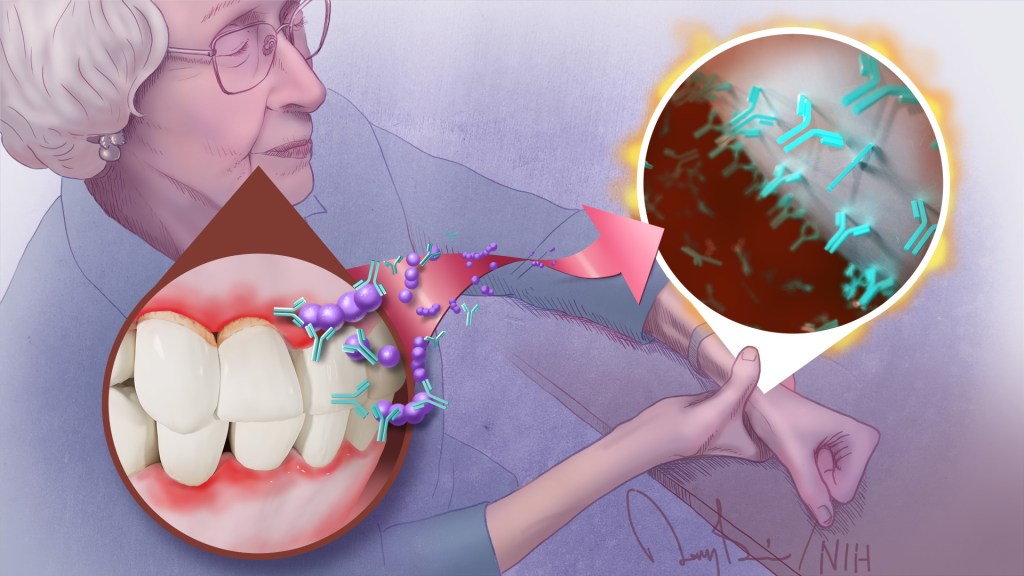
To keep your teeth and gums healthy for a lifetime, it’s important to brush and floss each day and see your dentist regularly. But what you might not often stop to consider is how essential good oral health really is to your overall well-being. The mouth, after all, is connected to the rest of the body, and oral infections can contribute to problems elsewhere.
A good case in point comes from a study just published in the journal Science Translational Medicine. The study, though small, offers some of the most convincing evidence yet for a direct link between gum, or periodontal, disease and the rheumatoid arthritis that flares most commonly in the hands, wrists, and knees [1]. If confirmed in larger follow-up studies, the finding suggests that one way for people with both diseases to contend with painful arthritic flare-ups will be to prevent them by practicing good oral hygiene and controlling their periodontal disease.
For many years, there had been suggestions that the oral bacteria causing periodontal disease might contribute to rheumatoid arthritis. For instance, past studies have found that periodontal disease occurs even more often in people with rheumatoid arthritis. People with both conditions also tend to have more severe arthritic symptoms that can be more stubbornly resistant to treatment.
What’s been missing is the precise underlying mechanisms to confirm the connection. To help connect the dots, a research team, which included Dana Orange, Rockefeller University, New York, NY, and William Robinson, Stanford University, Stanford, CA, decided to look closer.
They looked first in the blood, not directly at an arthritic joint or an inflamed periodontium, the tissues that hold a tooth in place. They were interested in whether telltale changes in the blood of people with rheumatoid arthritis correlated with the start of another painful flare-up in one or more of their joints.
One of those possible changes involves proteins that carry a particular chemical modification that places the amino acid citrulline on their surface. These citrulline-marked proteins are found in many parts of the human body, including the joints. Intriguingly, they also are present on bacteria, including those in the mouth.
Because of this bacterial connection, the researchers looked in the blood for a specific set of antibodies known as ACPAs, short for anti-citrullinated protein antibodies. They recognize citrullinated proteins that are foreign to the body and mark them for attack.
But the attack isn’t always perfectly aimed, and studies have shown the presence of ACPAs in the joints of people with rheumatoid arthritis is associated with increasing disease activity and more frequent arthritis flares. Periodontal disease, too, is especially common in people with rheumatoid arthritis who have abnormally high levels of circulating ACPAs.
In the new study, the researchers followed five women with rheumatoid arthritis for one to four years. Two of them had severe periodontal disease while the other three had no periodontal disease.
Each week, the study volunteers provided a small blood sample for researchers to study changes at the level of RNA, the genetic material that encodes proteins. They also studied changes in certain immune cells, along with any changes in their medication, dental care, or arthritis symptoms. For additional information, they also looked at blood and joint fluid samples from 67 other people with and without arthritis, including individuals with healthy gums or mild, moderate, or severe periodontal disease.
Overall, the evidence shows that people with more severe periodontal disease experienced repeated influxes of oral bacteria into their blood even when they hadn’t had a recent dental procedure. These findings suggested that when their inflamed gums became more damaged and “leaky,” bacteria in the mouth could spill into the bloodstream.
The researchers also found that those oral invaders carried many citrullinated proteins. Once they got into the bloodstream, inflammatory immune cells detected them and released ACPAs.
The researchers showed in the lab that those antibodies bind the same oral bacteria detected in the blood of people with periodontal disease and rheumatoid arthritis. In fact, those with both conditions had a wide variety of genetically distinct ACPAs, as would be expected if their immune systems were challenged repeatedly over time with oral bacteria.
The overarching idea is that these antibodies prime the immune system to attack oral bacteria. But after it gets started, the attack mistakenly expands and targets citrullinated proteins in the joints. That triggers a flare-up in a joint and the characteristic inflammation, stiffness, and joint damage.
While more study is needed to fill in the molecular details, this discovery raises an encouraging possibility. Taking care of your teeth and periodontal disease isn’t just a wise idea to maintain good oral health over a lifetime. For some of the approximately 1 million Americans with rheumatoid arthritis, it may help to manage and perhaps even prevent a painful flare-up in one or more of their affected joints.
Reference:
[1] Oral mucosal breaks trigger anti-citrullinated bacterial and human protein antibody responses in rheumatoid arthritis. Brewer RC, Lanz TV, Hale CR, Sepich-Poore GD, Martino C, Swafford AD, Carroll TS, Kongpachith S, Blum LK, Elliott SE, Blachere NE, Parveen S, Fak J, Yao V, Troyanskaya O, Frank MO, Bloom MS, Jahanbani S, Gomez AM, Iyer R, Ramadoss NS, Sharpe O, Chandrasekaran S, Kelmenson LB, Wang Q, Wong H, Torres HL, Wiesen M, Graves DT, Deane KD, Holers VM, Knight R, Darnell RB, Robinson WH, Orange DE. Sci Transl Med. 2023 Feb 22;15(684):eabq8476.
Links:
Rheumatoid Arthritis (National Institute of Arthritis and Musculoskeletal and Skin Diseases)
Periodontal (Gum) Disease (National Institute of Dental and Craniofacial Research/NIH)
Oral Hygiene (NIDCR)
Dana Orange (Rockefeller University, New York NY)
Robinson Lab (Stanford University, Stanford, CA)
NIH Support: National Institute of Arthritis and Musculoskeletal and Skin Diseases; National Institute of Allergy and Infectious Diseases; National Human Genome Research Institute; National Institute of General Medical Sciences; National Center for Advancing Translational Sciences; National Cancer Institute
Exploring Drug Repurposing for COVID-19 Treatment
Posted on by Dr. Francis Collins

It usually takes more than a decade to develop a safe, effective anti-viral therapy. But, when it comes to coronavirus disease 2019 (COVID-19), we don’t have that kind of time. One way to speed the process may be to put some old drugs to work against this new disease threat. This is generally referred to as “drug repurposing.”
NIH has been doing everything possible to encourage screens of existing drugs that have been shown safe for human use. In a recent NIH-funded study in the journal Nature, researchers screened a chemical “library” that contained nearly 12,000 existing drug compounds for their potential activity against SARS-CoV-2, the novel coronavirus that causes COVID-19 [1]. The results? In tests in both non-human primate and human cell lines grown in laboratory conditions, 21 of these existing drugs showed potential for repurposing to thwart the novel coronavirus—13 of them at doses that likely could be safely given to people. The majority of these drugs have been tested in clinical trials for use in HIV, autoimmune diseases, osteoporosis, and other conditions.
These latest findings come from an international team led by Sumit Chanda, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA. The researchers took advantage of a small-molecule drug library called ReFRAME [2], which was created in 2018 by Calibr, a non-profit drug discovery division of Scripps Research, La Jolla, CA.
In collaboration with Yuen Kwok-Yung’s team at the University of Hong Kong, the researchers first developed a high-throughput method that enabled them to screen rapidly each of the 11,987 drug compounds in the ReFRAME library for their potential to block SARS-CoV-2 in cells grown in the lab. The first round of testing narrowed the list of possible COVID-19 drugs to about 300. Next, using lower concentrations of the drugs in cells exposed to a second strain of SARS-CoV-2, they further narrowed the list to 100 compounds that could reliably limit growth of the coronavirus by at least 40 percent.
Generally speaking, an effective anti-viral drug is expected to show greater activity as its concentration is increased. So, Chanda’s team then tested those 100 drugs for evidence of such a dose-response relationship. Twenty-one of them passed this test. This group included remdesivir, a drug originally developed for Ebola virus disease and recently authorized by the U.S. Food and Drug Administration (FDA) for emergency use in the treatment of COVID-19. Remdesivir could now be considered a positive control.
These findings raised another intriguing question: Could any of the other drugs with a dose-response relationship work well in combination with remdesivir to block SARS-CoV-2 infection? Indeed, the researchers found that four of them could.
Further study showed that some of the most promising drugs on the list reduced the number of SARS-CoV-2 infected cells by 65 to 85 percent. The most potent of these was apilimod, a drug that has been evaluated in clinical trials for treating Crohn’s disease, rheumatoid arthritis, and other autoimmune conditions. Apilimod is now being evaluated in the clinic for its ability to prevent the progression of COVID-19. Another potential antiviral to emerge from the study is clofazimine, a 70-year old FDA-approved drug that is on the World Health Organization’s list of essential medicines for the treatment of leprosy.
Overall, the findings suggest that there may be quite a few existing drugs and/or experimental drugs fairly far along in the development pipeline that have potential to be repurposed for treating COVID-19. What’s more, some of them might also work well in combination with remdesivir, or perhaps other drugs, as treatment “cocktails,” such as those used to successfully treat HIV and hepatitis C.
This is just one of a wide variety of drug screening efforts that are underway, using different libraries and different assays to detect activity against SARS-CoV-2. The NIH’s National Center for Advancing Translational Sciences has established an open data portal to collect all of these data as quickly and openly as possible. As NIH continues its efforts to use the power of science to end the COVID-19 pandemic, it is critically important that we explore as many avenues as possible for developing diagnostics, treatments, and vaccines.
References:
[1] Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Riva L, Yuan S, Yin X, et al. Nature. 2020 Jul 24 [published online ahead of print]
[2] The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Janes J, Young ME, Chen E, et al. Proc Natl Acad Sci USA. 2018;115(42):10750-10755.
Links:
Coronavirus (COVID-19) (NIH)
ReFRAMEdb (Scripps Research, La Jolla, CA)
The Chanda Lab (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA)
Yuen Kwok-Yung (University of Hong Kong)
OpenData|Covid-19 (National Center for Advancing Translational Sciences/NIH)
NIH Support: National Institute of Allergy and Infectious Diseases; National Institute of General Medical Sciences
A New Tool in the Toolbox: New Method Traces Free-Floating DNA Back to Its Source
Posted on by Dr. Francis Collins

Caption: DNA (blue) loops around nucleosomes (gray) and is bound by transcription factors (red), proteins that switch genes on and off and act in a tissue-specific manner. When cells die, enzymes (scissors) chop up areas between the nucleosomes and transcription factors, releasing DNA fragments in unique patterns. By gathering the released DNA fragments in blood, researchers can tell which types of cells produced them.
Credit: Shendure Lab/University of Washington
When cells die, scissor-like enzymes snip their DNA into tiny fragments that leak into the bloodstream and other bodily fluids. Researchers have been busy in recent years working on ways to collect these free-floating bits of DNA and explore their potential use in clinical care.
These approaches, sometimes referred to as “liquid biopsies,” hinge on the ability to distinguish specific DNA fragments from the body’s normal background of “cell-free” DNA, most of which comes from dying white blood cells. Seeking other sources for cell-free DNA in particular situations is beginning to bear fruit, however. Current applications include: 1) a test in maternal blood to look for DNA from the fetus (actually from the fetal component of the placenta), which provides a means of detecting a possible genetic abnormality; 2) a test in a cancer patient’s blood to look for cancer-specific mutations, as a way of assessing response to treatment or early signs of relapse; and 3) a test in an organ transplant recipient, where increasing abundance of DNA fragments from the donor can be an early sign of rejection.
But recent proposals have been floated about looking for cell-free DNA in healthy individuals, as an early sign of some health problems. Suppose something was found—how could you know the source? Now a team of NIH-funded researchers has devised a new method that uses distinctive features of DNA packaging to provide an additional layer of information about the origins of free-floating DNA, vastly expanding the potential uses for such tests [1].
Meet Alex—Before and After NIH Clinical Trial
Posted on by Dr. Francis Collins
Alex Barton recently turned 17. That’s incredible because Alex was born with a rare, often fatal genetic disease and wasn’t expected to reach his teenage years.
When Alex was born, he looked like he’d been dipped in boiling water: his skin was bright red and blistered. He spent most of his time sleeping. When awake, he screamed in agony from headaches, joint pain, and rashes. After a torturous 14 months, a rheumatologist told his mother that Alex suffered from Neonatal-Onset Multisystem Inflammatory Disease (NOMID). The doctor showed her a brief and scary paragraph in a medical text. Kate Barton, Alex’s mother, admitted that it “knocked her over like a freight train.”
NIH Research Leads to New Rheumatoid Arthritis Drug
Posted on by Dr. Francis Collins

Copyright (2012) American College of Rheumatology.
About 1.5 million [1] people in the US suffer from rheumatoid arthritis (RA). It is a chronic illness in which the immune system, which protects us from viral and bacterial invaders, turns on our own body and viciously attacks the membranes that line our joints. The consequences can be excruciating: pain, swelling, stiffness, and decreased mobility. Over time, the joints can become permanently contorted, as in this X-ray image.
There are several RA medications on the market, but I want to tell you about a new one called tofacitinib, a pill which the FDA approved late last year [2]. The drug works by targeting a protein called Janus kinase 3, which was discovered by John O’Shea and colleagues here at the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) 20 years ago [3]. As I mentioned in a previous post it takes a really long time to go from a basic discovery to a drug—in most cases nearly 15 years. This drug has been even longer in the making! Shortly after discovering Janus kinase 3 in 1993, NIAMS researchers also revealed its role in inflammation, leading to a public-private collaboration with Pfizer that has now culminated in the approval of tofacitinib.



