cancer treatment
Microbe Normally Found in the Mouth May Drive Progression of Colorectal Cancer
Posted on by Dr. Monica M. Bertagnolli

Colorectal cancer is a leading cause of death from cancer in the United States. We know that risk of colorectal cancer goes up with age, certain coexisting health conditions, family history, smoking, alcohol use, and other factors. Researchers are also trying to learn more about what leads colorectal cancer to grow and spread. Now, findings from a new study supported in part by NIH add to evidence that colorectal tumor growth may be driven by a surprising bad actor: a microbe that’s normally found in the mouth.1
The findings, reported in Nature, suggest that a subtype of the bacterium Fusobacterium nucleatum has distinct genetic properties that may allow it to withstand acidic conditions in the stomach, infect colorectal tumors, and potentially drive their growth, which may lead to poorer patient outcomes. The discoveries suggest that the microbe could eventually be used as a target for detecting and treating colorectal cancer.
The study was conducted by a team led by Susan Bullman and Christopher D. Johnston at the Fred Hutchinson Cancer Center in Seattle. In 2022, the team published findings from a pair of studies implicating Fusobacterium nucleatum in the progression and spread of colorectal cancer.2,3 Their findings weren’t the first to suggest a link between the microbe and colorectal cancer. But their work offered important evidence that the microbe might alter colorectal tumors in ways that made them more likely to grow and spread. They also found that the microbe may affect the way colorectal cancer responds to or resists chemotherapy treatment.
Follow-up studies suggested there might be more to the story, pointing to the possibility that certain strains of the bacterium might differ from others in important ways. The findings suggested that there may be a more specific subtype, not yet defined, that was responsible for driving colorectal cancer growth.
To look deeper into this in the new study, Bullman and Johnston, with first author Martha Zepeda Rivera, analyzed a collection of 55 strains of the microbe taken from human colorectal cancer samples. They also compared these at the genetic level to another 80 strains of the microbe taken from the mouths of people who didn’t have cancer.
Their studies uncovered 483 genetic factors that turned up more often in Fusobacterium nucleatum from colorectal tumors. Those strains mainly belonged to a subspecies called Fusobacterium nucleatum animalis (Fna). More detailed study led to another surprise. The Fna included two genetically distinct groups or “clades” that had never been described, which the researchers called Fna C1 and C2. It turned out that only Fna C2 occurs at high levels in colorectal tumors.
The researchers found that this specific subtype within colorectal tumors carries 195 genetic factors that may allow it to grow more rapidly, withstand the acidic environment in the stomach, and take up residence in the gastrointestinal tract, where it can drive colorectal cancer growth. When the researchers infected a mouse model of colitis, a condition involving inflamed intestines that is a risk factor for colorectal cancer, they found that Fna C2 caused the development of more tumors compared to those infected with Fna C1.
Studies of tumors from 116 patients with colorectal cancer also showed more Fna C2. It was elevated in about 50% of cases. In fact, only this strain turned up more often in cancer compared to healthy tissue nearby. Stool samples of 627 people with colorectal cancer and 619 healthy people also showed more of this specific microbial strain in association with cancer.
This discovery is important because it suggests it’s only the Fna C2 subtype that’s associated with driving colorectal tumor growth, meaning it could help in the development of new methods for colorectal cancer screening and treatment. The researchers suggest it may one day even be possible to develop microbial-based therapies using modified versions of the bacterial strain to deliver treatments straight into tumors.
In addition, while the microbe is normally found in healthy mouths, it’s also enriched in periodontal (gum) disease, dental infections, and oral cancers.4 It will be interesting to learn more in future studies about the connections between various Fusobacterium nucleatum subtypes, oral health, and other health conditions throughout the body, including colorectal cancer.
References:
[1] Zepeda-Rivera M, et al. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature. DOI: 10.1038/s41586-024-07182-w (2024).
[2] LaCourse KD, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. DOI: 10.1016/j.celrep.2022.111625 (2022).
[3] Galeano Niño JL, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. DOI: 10.1038/s41586-022-05435-0 (2022).
[4] Chen Y, et al. More Than Just a Periodontal Pathogen –the Research Progress on Fusobacterium nucleatum. Front Cell Infect Microbiol. DOI: 10.3389/fcimb.2022.815318 (2022).
NIH Support: National Institute of Dental and Craniofacial Research, National Cancer Institute
Immune Checkpoint Discovery Has Implications for Treating Cancer and Autoimmune Diseases
Posted on by Dr. Monica M. Bertagnolli
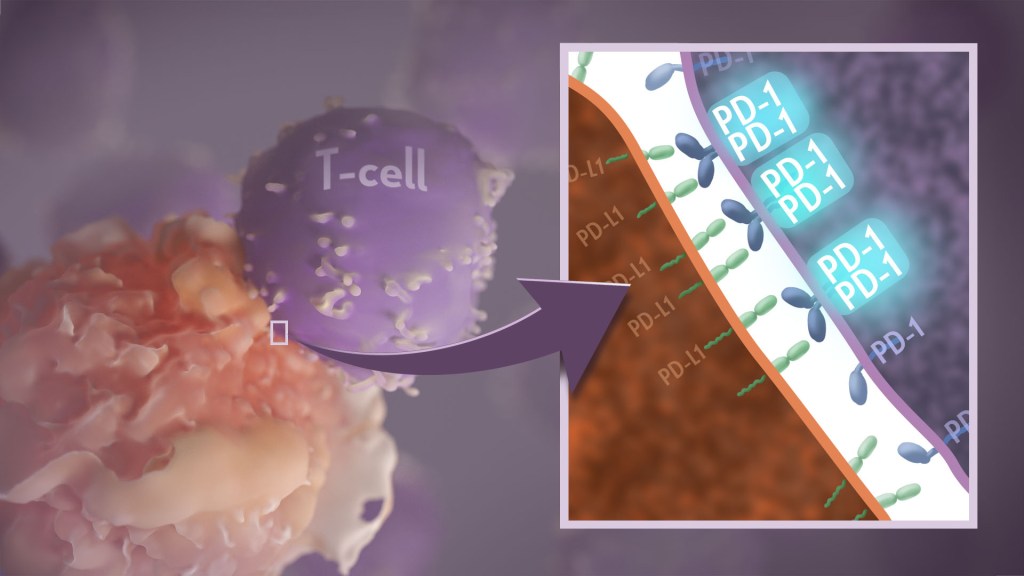
Your immune system should ideally recognize and attack infectious invaders and cancerous cells. But the system requires safety mechanisms, or brakes, to keep it from damaging healthy cells. To do this, T cells—the immune system’s most powerful attackers—rely on immune “checkpoints” to turn immune activation down when they receive the right signal. While these interactions have been well studied, a research team supported in part by NIH has made an unexpected discovery into how a key immune checkpoint works, with potentially important implications for therapies designed to boost or dampen immune activity to treat cancer and autoimmune diseases.1
The checkpoint in question is a protein called programmed cell death-1 (PD-1). Here’s how it works: PD-1 is a receptor on the surface of T cells, where it latches onto certain proteins, known as PD-L1 and PD-L2, on the surface of other cells in the body. When this interaction occurs, a signal is sent to the T cells that stops them from attacking these other cells.
Cancer cells often take advantage of this braking system, producing copious amounts of PD-L1 on their surface, allowing them to hide from T cells. An effective class of immunotherapy drugs used to treat many cancers works by blocking the interaction between PD-1 and PD-L1, to effectively release the brakes on the immune system to allow the T cells to unleash an assault on cancer cells. Researchers have also developed potential treatments for autoimmune diseases that take the opposite tact: stimulating PD-1 interaction to keep T cells inactive. These PD-1 “agonists” have shown promise in clinical trials as treatments for certain autoimmune diseases.
However, to fulfill the promise of these approaches for treating cancer and autoimmune diseases, a better understanding of precisely how PD-1 works to suppress T cell activity is still needed. The thinking has long been that individual PD-1 receptors act alone. But, as reported in Science Immunology, it turns out that this may not usually be the case. A team led by Jun Wang and Xiangpeng Kong of New York University Langone Health’s Perlmutter Cancer Center, with Elliot Philips of NYU and Michael Dustin of the University of Oxford, U.K., used sophisticated techniques to look for evidence of what happens when PD-1 proteins work together in pairs.
They found that PD-1’s tendency to link, or not link, with a second PD-1 protein to form what’s known as a “dimer” depends on interactions with portions of the protein that cross the immune cell membrane. They also found that, when PD-1 receptors pair up, they do a better job of squashing immune responses. The findings also showed that a single change in the amino acid structure in the portion of PD-1 that crosses the cell membrane can strengthen or weaken immune responses.
One reason why these fundamental discoveries are exciting is they suggest that interfering with PD-1’s ability to form dimers might make immunotherapy treatments for cancer more effective. In addition, treatments that strengthen interactions between paired PD-1 receptors might aid in the design of promising new drug classes that are intended to tamp down inflammation seen in people with some autoimmune diseases, including rheumatoid arthritis, lupus, and type 1 diabetes. The research team now plans to conduct further investigations of PD-1 blockers and agonists to explore whether these findings could eventually lead to more effective treatments for both cancer and autoimmune diseases.
Reference:
[1] Philips EA, et al. Transmembrane domain-driven PD-1 dimers mediate T cell inhibition. Science Immunology. DOI: 10.1126/sciimmunol.ade6256 (2024).
NIH Support: National Institute of Allergy and Infectious Diseases, National Cancer Institute, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institute of General Medical Sciences
Rice-Sized Device Tests Brain Tumor’s Drug Responses During Surgery
Posted on by Lawrence Tabak, D.D.S., Ph.D.

Scientists have made remarkable progress in understanding the underlying changes that make cancer grow and have applied this knowledge to develop and guide targeted treatment approaches to vastly improve outcomes for people with many cancer types. And yet treatment progress for people with brain tumors known as gliomas—including the most aggressive glioblastomas—has remained slow. One reason is that doctors lack tests that reliably predict which among many therapeutic options will work best for a given tumor.
Now an NIH-funded team has developed a miniature device with the potential to change this for the approximately 25,000 people diagnosed with brain cancers in the U.S. each year [1]. When implanted into cancerous brain tissue during surgery, the rice-sized drug-releasing device can simultaneously conduct experiments to measure a tumor’s response to more than a dozen drugs or drug combinations. What’s more, a small clinical trial reported in Science Translational Medicine offers the first evidence in people with gliomas that these devices can safely offer unprecedented insight into tumor-specific drug responses [2].
These latest findings come from a Brigham and Women’s Hospital, Boston, team led by Pierpaolo Peruzzi and Oliver Jonas. They recognized that drug-screening studies conducted in cells or tissue samples in the lab too often failed to match what happens in people with gliomas undergoing cancer treatment. Wide variation within individual brain tumors also makes it hard to predict a tumor’s likely response to various treatment options.
It led them to an intriguing idea: Why not test various therapeutic options in each patient’s tumor? To do it, they developed a device, about six millimeters long, that can be inserted into a brain tumor during surgery to deliver tiny doses of up to 20 drugs. Doctors can then remove and examine the drug-exposed cancerous tissue in the laboratory to determine each treatment’s effects. The data can then be used to guide subsequent treatment decisions, according to the researchers.
In the current study, the researchers tested their device on six study volunteers undergoing brain surgery to remove a glioma tumor. For each volunteer, the device was implanted into the tumor and remained in place for about two to three hours while surgeons worked to remove most of the tumor. Next, the device was taken out along with the last piece of a tumor at the end of the surgery for further study of drug responses.
Importantly, none of the study participants experienced any adverse effects from the device. Using the devices, the researchers collected valuable data, including how a tumor’s response changed with varying drug concentrations or how each treatment led to molecular changes in the cancerous cells.
More research is needed to better understand how use of such a device might change treatment and patient outcomes in the longer term. The researchers note that it would take more than a couple of hours to determine how treatments produce less immediate changes, such as immune responses. As such, they’re now conducting a follow-up trial to test a possible two-stage procedure, in which their device is inserted first using minimally invasive surgery 72 hours prior to a planned surgery, allowing longer exposure of tumor tissue to drugs prior to a tumor’s surgical removal.
Many questions remain as they continue to optimize this approach. However, it’s clear that such a device gives new meaning to personalized cancer treatment, with great potential to improve outcomes for people living with hard-to-treat gliomas.
References:
[1] National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Brain and Other Nervous System Cancer.
[2] Peruzzi P et al. Intratumoral drug-releasing microdevices allow in situ high-throughput pharmaco phenotyping in patients with gliomas. Science Translational Medicine DOI: 10.1126/scitranslmed.adi0069 (2023).
Links:
Brain Tumors – Patient Version (National Cancer Institute/NIH)
Pierpaolo Peruzzi (Brigham and Women’s Hospital, Boston, MA)
Jonas Lab (Brigham and Women’s Hospital, Boston, MA)
NIH Support: National Cancer Institute, National Institute of Biomedical Imaging and Bioengineering, National Institute of Neurological Disorders and Stroke
New 3D Atlas of Colorectal Cancer Promises Improved Diagnosis, Treatment
Posted on by Lawrence Tabak, D.D.S., Ph.D.

This year, too many Americans will go to the doctor for tissue biopsies to find out if they have cancer. Highly trained pathologists will examine the biopsies under a microscope for unusual cells that show the telltale physical features of a suspected cancer. As informative as the pathology will be for considering the road ahead, it would be even more helpful if pathologists had the tools to look widely inside cells for the actual molecules giving rise to the tumor.
Working this “molecular information” into the pathology report would bring greater diagnostic precision, drilling down to the actual biology driving the growth of the tumor. It also would help doctors to match the right treatments to a patient’s tumor and not waste time on drugs that will be ineffective.
That’s why researchers have been busy building the needed tools and also mapping out molecular atlases of common cancers. These atlases, really a series of 3D spatial maps detailing various biological features within the tumor, keep getting better all the time. That includes the comprehensive atlas of colorectal cancer just published in the journal Cell [1].
This colorectal atlas comes from an NIH-supported team led by Sandro Santagata, Brigham and Women’s Hospital, Boston, and Peter Sorger, Harvard Medical School, Cambridge, MA, in collaboration with investigators at Vanderbilt University, Nashville, TN. The colorectal atlas joins their previously published high-definition map of melanoma [2], and both are part of the Human Tumor Atlas Network that’s supported by NIH’s National Cancer Institute.
What’s so interesting with the colorectal atlas is the team combined traditional pathology with a sophisticated technique for imaging single cells, enabling them to capture their fine molecular details in an unprecedented way.
They did it using a cutting-edge technique known as cyclic immunofluorescence, or CyCIF. In CyCIF, researchers use many rounds of highly detailed molecular imaging on each tissue sample to generate a rich collection of molecular-level data, cell by cell. Altogether, the researchers captured this fine-scale visual information for nearly 100 million cancer cells isolated from tumor samples representing 93 individuals diagnosed with colorectal cancer.
With this single-cell information in hand, they next created detailed 2D maps covering the length and breadth of large portions of the colorectal cancers under study. Finally, with the aid of first author Jia-Ren Lin, also at Harvard Medical School, and colleagues they stitched together their 2D maps to produce detailed 3D reconstructions showing the length, breadth, and height of the tumors.
This more detailed view of colorectal cancer has allowed the team to explore differences between normal and tumor tissues, as well as variations within an individual tumor. In fact, they’ve uncovered physical features that had never been discovered.
For instance, an individual tumor has regions populated with malignant cells, while other areas look less affected by the cancer. In between are transitional areas that correspond to molecular gradients of information. With this high-resolution map as their guide, researchers can now study what this all might mean for the diagnosis, treatment, and prognosis of colorectal cancer.
The atlas also shows that the presence of immune cells varies dramatically within a single tumor. That’s an important discovery because of its potential implications for immunotherapies, in which treatments aim to unleash the immune system in the fight against cancer.
The maps also provide new insights into tumor structure. For example, scientists had previously identified what they thought were 2D pools of a mucus-like substance called mucin with clusters of cancer cells suspended inside. However, the new 3D reconstruction make clear that these aren’t simple mucin pools. Rather, they are cross sections of larger intricate caverns of mucin interconnected by channels, into which cancer cells make finger-like projections.
The good news is the researchers already are helping to bring these methods into the cancer clinic. They also hope to train other scientists to build their own cancer atlases and grow the collection even more.
In the meantime, the team will refine its 3D tumor reconstructions by integrating new imaging technologies and even more data into their maps. It also will map many more colorectal cancer samples to capture the diversity of their basic biology. Also of note, having created atlases for melanoma and colorectal cancer, the team has plans to tackle breast and brain cancers next.
Let me close by saying, if you’re between the ages of 45 and 75, don’t forget to stay up to date on your colorectal cancer screenings. These tests are very good, and they could save your life.
References:
[1] Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer. Lin JR, Wang S, Coy S, Chen YA, Yapp C, Tyler M, Nariya MK, Heiser CN, Lau KS, Santagata S, Sorger PK. Cell. 2023 Jan 19;186(2):363-381.e19.
[2] The spatial landscape of progression and immunoediting in primary melanoma at single-cell resolution. Nirmal AJ, Maliga Z, Vallius T, Quattrochi B, Chen AA, Jacobson CA, Pelletier RJ, Yapp C, Arias-Camison R, Chen YA, Lian CG, Murphy GF, Santagata S, Sorger PK. Cancer Discov. 2022 Jun 2;12(6):1518-1541.
Links:
Colorectal Cancer (National Cancer Institute/NIH)
Human Tumor Atlas Network (NCI)
CyCIF-Cyclic Immunofluorescence (Harvard Medical School, Cambridge, MA)
Sandro Santagata (Brigham and Women’s Hospital, Boston)
Peter Sorger (Harvard Medical School)
Jia-Ren Lin (Harvard Medical School)
NIH Support: National Cancer Institute; National Institute of General Medical Sciences; National Institute of Diabetes and Digestive and Kidney Diseases
Precision Oncology: Gene Changes Predict Immunotherapy Response
Posted on by Dr. Francis Collins

Caption: Adapted from scanning electron micrograph of cytotoxic T cells (red) attacking a cancer cell (white).
Credits: Rita Elena Serda, Baylor College of Medicine; Jill George, NIH
There’s been tremendous excitement in the cancer community recently about the life-saving potential of immunotherapy. In this treatment strategy, a patient’s own immune system is enlisted to control and, in some cases, even cure the cancer. But despite many dramatic stories of response, immunotherapy doesn’t work for everyone. A major challenge has been figuring out how to identify with greater precision which patients are most likely to benefit from this new approach, and how to use that information to develop strategies to expand immunotherapy’s potential.
A couple of years ago, I wrote about early progress on this front, highlighting a small study in which NIH-funded researchers were able to predict which people with colorectal and other types of cancer would benefit from an immunotherapy drug called pembrolizumab (Keytruda®). The key seemed to be that tumors with defects affecting the “mismatch repair” pathway were more likely to benefit. Mismatch repair is involved in fixing small glitches that occur when DNA is copied during cell division. If a tumor is deficient in mismatch repair, it contains many more DNA mutations than other tumors—and, as it turns out, immunotherapy appears to be most effective against tumors with many mutations.
Now, I’m pleased to report more promising news from that clinical trial of pembrolizumab, which was expanded to include 86 adults with 12 different types of mismatch repair-deficient cancers that had been previously treated with at least one type of standard therapy [1]. After a year of biweekly infusions, more than half of the patients had their tumors shrink by at least 30 percent—and, even better, 18 had their tumors completely disappear!

