DNA editing
Experts Conclude Heritable Human Genome Editing Not Ready for Clinical Applications
Posted on by Dr. Francis Collins

We stand at a critical juncture in the history of science. CRISPR and other innovative genome editing systems have given researchers the ability to make very precise changes in the sequence, or spelling, of the human DNA instruction book. If these tools are used to make non-heritable edits in only relevant tissues, they hold enormous potential to treat or even cure a wide range of devastating disorders, such as sickle cell disease, inherited neurologic conditions, and muscular dystrophy. But profound safety, ethical, and philosophical concerns surround the use of such technologies to make heritable changes in the human genome—changes that can be passed on to offspring and have consequences for future generations of humankind.
Such concerns are not hypothetical. Two years ago, a researcher in China took it upon himself to cross this ethical red line and conduct heritable genome editing experiments in human embryos with the aim of protecting the resulting babies against HIV infection. The medical justification was indefensible, the safety issues were inadequately considered, and the consent process was woefully inadequate. In response to this epic scientific calamity, NIH supported a call by prominent scientists for an international moratorium on human heritable, or germline, genome editing for clinical purposes.
Following on the heels of this unprecedented ethical breach, the U.S. National Academy of Sciences, U.S. National Academy of Medicine, and the U.K. Royal Society convened an international commission, sponsored by NIH, to conduct a comprehensive review of the clinical use of human germline genome editing. The 18-member panel, which represented 10 nations and four continents, included experts in genome editing technology; human genetics and genomics; psychology; reproductive, pediatric, and adult medicine; regulatory science; bioethics; and international law. Earlier this month, this commission issued its consensus study report, entitled Heritable Human Genome Editing [1].
The commission was designed to bring together thought leaders around the globe to engage in serious discussions about this highly controversial use of genome-editing technology. Among the concerns expressed by many of us was that if heritable genome editing were allowed to proceed without careful deliberation, the enormous potential of non-heritable genome editing for prevention and treatment of disease could become overshadowed by justifiable public outrage, fear, and disgust.
I’m gratified to say that in its new report, the expert panel closely examined the scientific and ethical issues, and concluded that heritable human genome editing is too technologically unreliable and unsafe to risk testing it for any clinical application in humans at the present time. The report cited the potential for unintended off-target DNA edits, which could have harmful health effects, such as cancer, later in life. Also noted was the risk of producing so-called mosaic embryos, in which the edits occur in only a subset of an embryo’s cells. This would make it very difficult for researchers to predict the clinical effects of heritable genome editing in human beings.
Among the many questions that the panel was asked to consider was: should society ever decide that heritable gene editing might be acceptable, what would be a viable framework for scientists, clinicians, and regulatory authorities to assess the potential clinical applications?
In response to that question, the experts replied: heritable gene editing, if ever permitted, should be limited initially to serious diseases that result from the mutation of one or both copies of a single gene. The first uses of these technologies should proceed incrementally and with extreme caution. Their potential medical benefits and harms should also be carefully evaluated before proceeding.
The commission went on to stress that before such an option could be on the table, all other viable reproductive possibilities to produce an embryo without a disease-causing alteration must be exhausted. That would essentially limit heritable gene editing to the exceedingly rare instance in which both parents have two copies of a recessive, disease-causing gene variant. Or another quite rare instance in which one parent has two copies of an altered gene for a dominant genetic disorder, such as Huntington’s disease.
Recognizing how unusual both scenarios would be, the commission held out the possibility that some would-be parents with less serious conditions might qualify if 25 percent or less of their embryos are free of the disease-causing gene variant. A possible example is familial hypercholesterolemia (FH), in which people carrying a mutation in the LDL receptor gene have unusually high levels of cholesterol in their blood. If both members of a couple are affected, only 25 percent of their biological children would be unaffected. FH can lead to early heart disease and death, but drug treatment is available and improving all the time, which makes this a less compelling example. Also, the commission again indicated that such individuals would need to have already traveled down all other possible reproductive avenues before considering heritable gene editing.
A thorny ethical question that was only briefly addressed in the commission’s report is the overall value to be attached to a couple’s desire to have a biological child. That desire is certainly understandable, although other options, such an adoption or in vitro fertilization with donor sperm, are available. This seems like a classic example of the tension between individual desires and societal concerns. Is the drive for a biological child in very high-risk situations such a compelling circumstance that it justifies asking society to start down a path towards modifying human germline DNA?
The commission recommended establishing an international scientific advisory board to monitor the rapidly evolving state of genome editing technologies. The board would serve as an access point for scientists, legislators, and the public to access credible information to weigh the latest progress against the concerns associated with clinical use of heritable human genome editing.
The National Academies/Royal Society report has been sent along to the World Health Organization (WHO), where it will serve as a resource for its expert advisory committee on human genome editing. The WHO committee is currently developing recommendations for appropriate governance mechanisms for both heritable and non-heritable human genome editing research and their clinical uses. That panel could issue its guidance later this year, which is sure to continue this very important conversation.
Reference:
[1] Heritable Human Genome Editing, Report Summary, National Academy of Sciences, September 2020.
Links:
“Heritable Genome Editing Not Yet Ready to Be Tried Safely and Effectively in Humans,” National Academies of Sciences, Engineering, and Medicine news release, Sep. 3, 2020.
International Commission on the Clinical Use of Human Germline Genome Editing (National Academies of Sciences, Engineering, and Medicine/Washington, D.C.)
Video: Report Release Webinar , International Commission on the Clinical Use of Human Germline Genome Editing (National Academies of Sciences, Engineering, and Medicine)
National Academy of Sciences (Washington, D.C.)
National Academy of Medicine (Washington, D.C.)
The Royal Society (London)
Find and Replace: DNA Editing Tool Shows Gene Therapy Promise
Posted on by Dr. Francis Collins
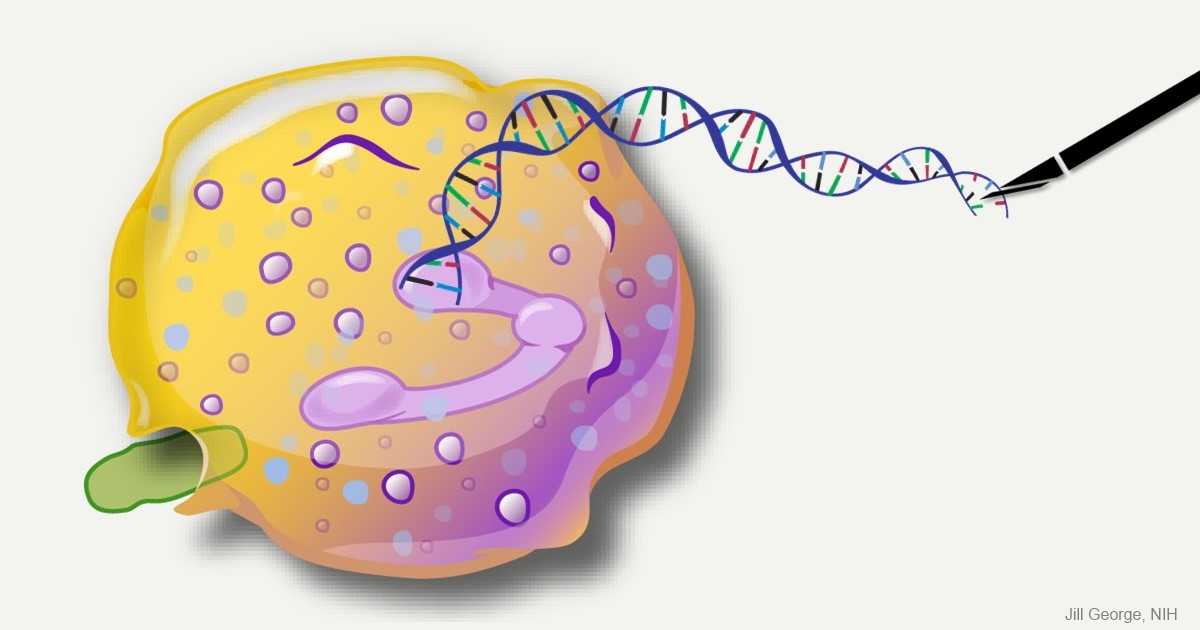
Caption: This image represents an infection-fighting cell called a neutrophil. In this artist’s rendering, the cell’s DNA is being “edited” to help restore its ability to fight bacterial invaders.
Credit: NIAID, NIH
For gene therapy research, the perennial challenge has been devising a reliable way to insert safely a working copy of a gene into relevant cells that can take over for a faulty one. But with the recent discovery of powerful gene editing tools, the landscape of opportunity is starting to change. Instead of threading the needle through the cell membrane with a bulky gene, researchers are starting to design ways to apply these tools in the nucleus—to edit out the disease-causing error in a gene and allow it to work correctly.
While the research is just getting under way, progress is already being made for a rare inherited immunodeficiency called chronic granulomatous disease (CGD). As published recently in Science Translational Medicine, a team of NIH researchers has shown with the help of the latest CRISPR/Cas9 gene-editing tools, they can correct a mutation in human blood-forming adult stem cells that triggers a common form of CGD. What’s more, they can do it without introducing any new and potentially disease-causing errors to the surrounding DNA sequence [1].
When those edited human cells were transplanted into mice, the cells correctly took up residence in the bone marrow and began producing fully functional white blood cells. The corrected cells persisted in the animal’s bone marrow and bloodstream for up to five months, providing proof of principle that this lifelong genetic condition and others like it could one day be cured without the risks and limitations of our current treatments.
Sickle Cell Disease: Gene-Editing Tools Point to Possible Ultimate Cure
Posted on by Dr. Francis Collins
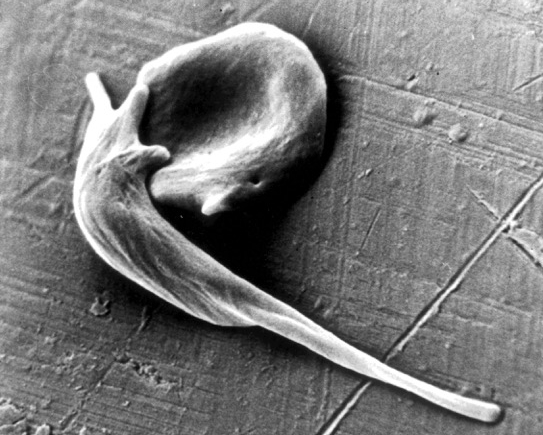
Caption: An electron micrograph showing two red blood cells deformed by crystalline hemoglobin into different “sickle” shapes characteristic of people with sickle cell disease.
Credit: Frans Kuypers: RBClab.com, UCSF Benioff Children’s Hospital Oakland
Scientists first described the sickle-shaped red blood cells that give sickle cell disease its name more than a century ago. By the 1950s, the precise molecular and genetic underpinnings of this painful and debilitating condition had become clear, making sickle cell the first “molecular disease” ever characterized. The cause is a single letter “typo” in the gene encoding oxygen-carrying hemoglobin. Red blood cells containing the defective hemoglobin become stiff, deformed, and prone to clumping. Individuals carrying one copy of the sickle mutation have sickle trait, and are generally fine. Those with two copies have sickle cell disease and face major medical challenges. Yet, despite all this progress in scientific understanding, nearly 70 years later, we still have no safe and reliable means for a cure.
Recent advances in CRISPR/Cas9 gene-editing tools, which the blog has highlighted in the past, have renewed hope that it might be possible to cure sickle cell disease by correcting DNA typos in just the right set of cells. Now, in a study published in Science Translational Medicine, an NIH-funded research team has taken an encouraging step toward this goal [1]. For the first time, the scientists showed that it’s possible to correct the hemoglobin mutation in blood-forming human stem cells, taken directly from donors, at a frequency that might be sufficient to help patients. In addition, their gene-edited human stem cells persisted for 16 weeks when transplanted into mice, suggesting that the treatment might also be long lasting or possibly even curative.
Creative Minds: Applying CRISPR Technology to Cancer Drug Resistance
Posted on by Dr. Francis Collins

Patrick Hsu
As a child, Patrick Hsu once settled a disagreement with his mother over antibacterial wipes by testing them in controlled experiments in the kitchen. When the family moved to Palo Alto, CA, instead of trying out for the football team or asking to borrow the family car like other high school kids might have done, Hsu went knocking on doors of scientists at Stanford University. He found his way into a neuroscience lab, where he gained experience with the fundamental tools of biology and a fascination for understanding how the brain works. But Hsu would soon become impatient with the tools that were available to ask some of the big questions he wanted to study.
As a Salk Helmsley Fellow and principal investigator at the Salk Institute for Biological Studies, La Jolla, CA, Hsu now works at the intersection of bioengineering, genomics, and neuroscience with a DNA editing tool called CRISPR/Cas9 that is revolutionizing the way scientists can ask and answer those big questions. (This blog has previously featured several examples of how this technology is revolutionizing biomedical research.) Hsu has received a 2015 NIH Director’s Early Independence award to adapt CRISPR/Cas9 technology so its use can be extended to that other critically important information-containing nucleic acid—RNA.Specifically, Hsu aims to develop ways to use this new tool to examine the role of a certain type of RNA in cancer drug resistance.
Popular Genome Editing Tool Gets Its Close-Up
Posted on by Dr. Francis Collins
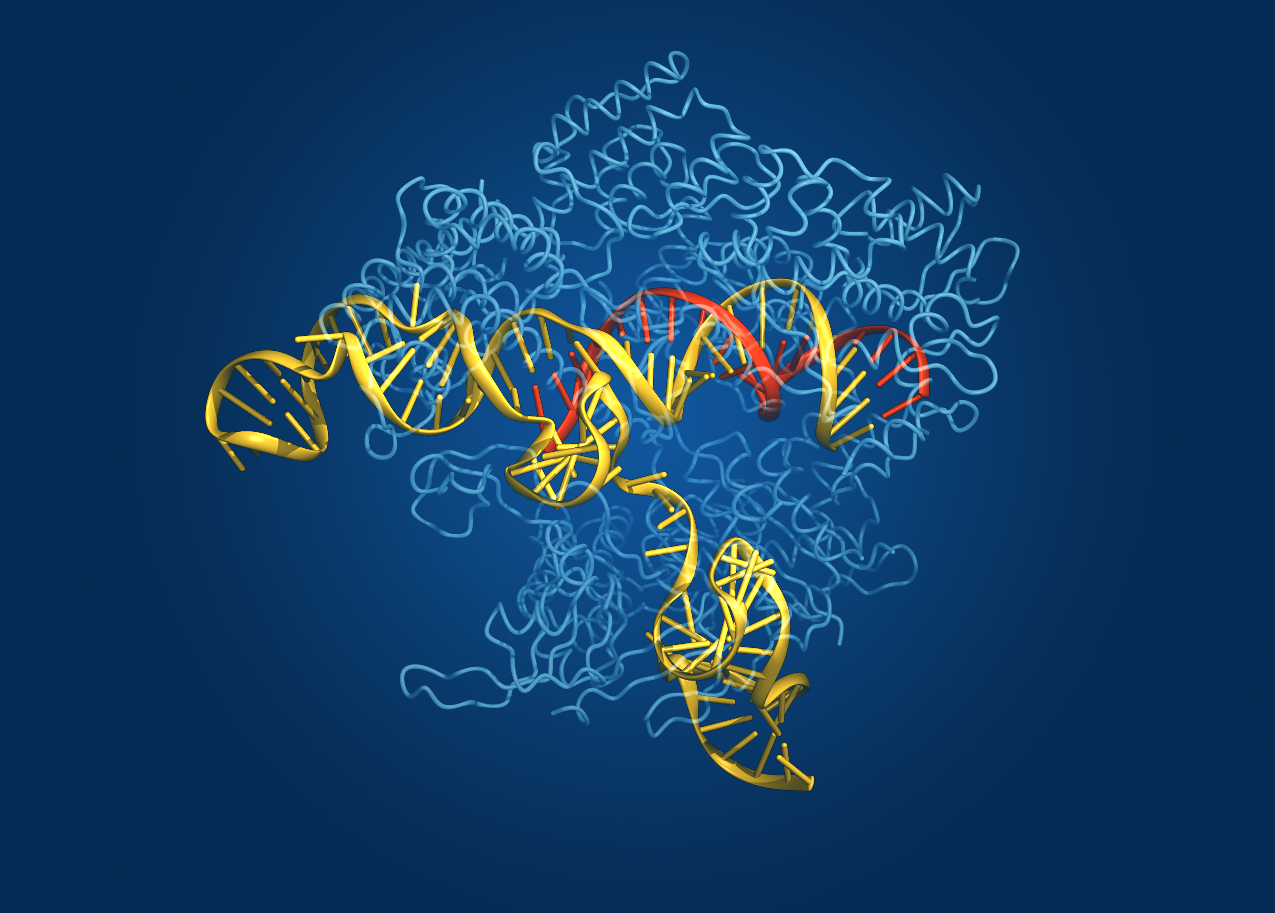
Caption: Crystal structure of the Cas9 gene-editing enzyme (light blue) in complex with an RNA guide (red) and its target DNA (yellow).
Credit: Bang Wong, Broad Institute of Harvard and MIT, Cambridge, MA
Exactly one hundred years ago, Max von Laue won the Nobel Prize in Physics for discovering that when a crystal is bombarded with X-rays, the beams bounce off the electrons surrounding the nucleus of each atom and scatter, interfering with each other (like ripples in a pond) and creating a unique pattern. These diffraction patterns could be used to decipher the arrangement of atoms in the crystal. Since then, X-ray crystallography has been used to chart a vast number of biological structures, including those of DNA, proteins, and even whole viruses.
Now, NIH-funded researchers at the Broad Institute of MIT and Harvard (Cambridge, MA) have teamed up with researchers at the University of Tokyo (Japan) to use crystallography to generate a high-definition map of an innovative tool for editing genomes. Their image reveals the structure of Cas9—an enzyme with an amazing ability to slice DNA with exquisite precision—in complex with a molecule of RNA that is guiding it to a targeted region of DNA [1].
The Cas9 enzyme was originally discovered in bacteria. It’s a key part of an ancient microbial immune system, called CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-Cas), that researchers recently discovered could be put to use as a tool for precisely altering DNA. This extraordinary system has been used to knock out genes in cells from bacteria, mice, and humans, and even to engineer monkeys with specific mutations that could serve as more accurate models of human disease.
Still, there’s room for improvement. Because Cas9 is rather large, Broad researcher Feng Zhang (a recipient of both the NIH Director’s Transformative Research Award and an NIH Director’s Pioneer Award) wants to trim the enzyme a bit so it could be packaged into viruses for new applications. Armed with the new crystal structure, Zhang’s team can now determine which regions of the enzyme are essential for editing DNA and which parts might be dispensable.
Another issue with Cas9 is that it occasionally makes errors, cutting the wrong region of DNA. Zhang thinks the new structural schematic of Cas9, along with the guide molecule RNA and target DNA, might point to ways in which the enzyme can be optimized to reduce the chance of errors.
Zhang’s team isn’t the only one interested in tweaking Cas9 to improve its engineering potential. A group led by Jennifer Doudna and Eva Nogales, both of the University of California, Berkeley, also recently used X-ray crystallography to generate images of two different versions of Cas9: one from Streptococcus pyogenes and the other from Actinomyces naeslundii [2]. By the way, the Foundation for the NIH recently named Doudna as the winner of its 2014 Lurie Prize in the Biomedical Sciences. The NIH grantee received the award for her pioneering role in the 2012 discovery of the CRISPR gene-editing technique.
Thanks to all of these new crystal structures, the scientific community is a step closer to realizing the full potential of Cas9/CRISPR technology to advance our understanding of disease and accelerate development of treatments and cures. So, here’s to our old friend crystallography and all of the exciting ways in which it will continue to expand our scientific horizons for years to come!
References:
[1] Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Cell. 2014 Feb 12. pii: S0092-8674(14)00156-1.
[2] Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Science. 2014 Feb 6
Links:
Zhang Lab, Broad Institute of Harvard and MIT, Cambridge, MA
Genome Engineering Resource Center maintained by the Zhang Lab
Nureki Lab, Department of Biophysics and Biochemistry, The University of Tokyo
Doudna Lab, University of California, Berkeley, CA
Foundation for the NIH to Award Lurie Prize in Biomedical Sciences to Jennifer Doudna from UC Berkeley, 25 February 2014
The Nogales Lab, University of California, Berkeley, CA
NIH Director’s Pioneer Award. (NIH Common Fund)
NIH Director’s Transformative Research Award. (NIH Common Fund)
NIH support: Office of the Director (Common Fund); National Institute of Mental Health; National Institute of General Medical Sciences
