neural progenitor cells
New Technology Opens Evolutionary Window into Brain Development
Posted on by Dr. Francis Collins

One of the great mysteries in biology is how we humans ended up with such large, complex brains. In search of clues, researchers have spent years studying the protein-coding genes activated during neurodevelopment. But some answers may also be hiding in non-coding regions of the human genome, where sequences called regulatory elements increase or decrease the activity of genes.
A fascinating example involves a type of regulatory element called a human accelerated region (HAR). Although “human” is part of this element’s name, it turns out that the genomes of all vertebrates—not just humans—contain the DNA segments now designated as HARs.
In most organisms, HARs show a relatively low rate of mutation, which means these regulatory elements have been highly conserved across species throughout evolutionary time [1]. The big exception is Homo sapiens, in which HARs have exhibited a much higher rate of mutations.
The accelerated rate of HARs mutations observed in humans suggest that, over the course of very long periods of time, these genomic changes might have provided our species with some sort of evolutionary advantage. What might that be? Many have speculated the advantage might involve the brain because HARs are often associated with genes involved in neurodevelopment. Now, in a paper published in the journal Neuron, an NIH-supported team confirms that’s indeed the case [2].
In the new work, researchers found that about half of the HARs in the human genome influence the activity, or expression, of protein-coding genes in neural cells and tissues during the brain’s development [3]. The researchers say their study—the most comprehensive to date of the 3,171 HARs in the human genome—firmly establishes that this type of regulatory element helps to drive patterns of neurodevelopmental gene activity specific to humans.
Yet to be determined is precisely how HARs affect the development of the human brain. The quest to uncover these details will no doubt shed new light on fundamental questions about the brain, its billions of neurons, and their trillions of interconnections. For example, why does human neural development span decades, longer than the life spans of most primates and other mammals? Answering such questions could also reveal new clues into a range of cognitive and behavioral disorders. In fact, early research has already made tentative links between HARs and neurodevelopmental conditions such as autism spectrum disorder and schizophrenia [3].
The latest work was led by Kelly Girskis, Andrew Stergachis, and Ellen DeGennaro, all of whom were in the lab of Christopher Walsh while working on the project. An NIH grantee, Walsh is director of the Allen Discovery Center for Brain Evolution at Boston Children’s Hospital and Harvard Medical School, which is supported by the Paul G. Allen Foundation Frontiers Group, and is an Investigator of the Howard Hughes Medical Institute.
Though HARs have been studied since 2006, one of the big challenges in systematically assessing them has been technological. The average length of a HAR is about 269 bases of DNA, but current technologies for assessing function can only easily analyze DNA molecules that span 150 bases or less.
Ryan Doan, who was then in the Walsh Lab, and his colleagues solved the problem by creating a new machine called CaptureMPRA. (MPRA is short for “massively parallel reporter assays.”) This technological advance cleverly barcodes HARs and, more importantly, makes it possible to analyze HARs up to about 500 bases in length.
Using CaptureMPRA technology in tandem with cell culture studies, researchers rolled up their sleeves and conducted comprehensive, full-sequence analyses of more than 3,000 HARs. In their initial studies, primarily in neural cells, they found nearly half of human HARs are active to drive gene expression in cell culture. Of those, 42 percent proved to have increased ability to enhance gene expression compared to their orthologues, or counterparts, in chimpanzees.
Next, the team integrated these data with an existing epigenetic dataset derived from developing human brain cells, as well as additional datasets generated from sorted brain cell types. They found that many HARs appeared to have the ability to increase the activity of protein-coding genes, while a smaller—but very significant—subset of the HARs appeared to be enhancing gene expression specifically in neural progenitor cells, which are responsible for making various neural cell types.
The data suggest that as the human HAR sequences mutated and diverged from other mammals, they increased their ability to enhance or sometimes suppress the activity of certain genes in neural cells. To illustrate this point, the researchers focused on two HARs that appear to interact specifically with a gene referred to as R17. This gene can have highly variable gene expression patterns not only in different human cell types, but also in cells from other vertebrates and non-vertebrates.
In the human cerebral cortex, the outermost part of the brain that’s responsible for complex behaviors, R17 is expressed only in neural progenitor cells and only at specific time points. The researchers found that R17 slows the progression of neural progenitor cells through the cell cycle. That might seem strange, given the billions of neurons that need to be made in the cortex. But it’s consistent with the biology. In the human, it takes more than 130 days for the cortex to complete development, compared to about seven days in the mouse.
Clearly, to learn more about how the human brain evolved, researchers will need to look for clues in many parts of the genome at once, including its non-coding regions. To help researchers navigate this challenging terrain, the Walsh team has created an online resource displaying their comprehensive HAR data. It will appear soon, under the name HAR Hub, on the University of California Santa Cruz Genome Browser.
References:
[1] An RNA gene expressed during cortical development evolved rapidly in humans. Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M Jr, Vanderhaeghen P, Haussler D. Nature. 2006 Sep 14;443(7108):167-72.
[2] Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Girskis KM, Stergachis AB, DeGennaro EM, Doan RN, Qian X, Johnson MB, Wang PP, Sejourne GM, Nagy MA, Pollina EA, Sousa AMM, Shin T, Kenny CJ, Scotellaro JL, Debo BM, Gonzalez DM, Rento LM, Yeh RC, Song JHT, Beaudin M, Fan J, Kharchenko PV, Sestan N, Greenberg ME, Walsh CA. Neuron. 2021 Aug 25:S0896-6273(21)00580-8.
[3] Mutations in human accelerated regions disrupt cognition and social behavior. Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, Gascon GG; Homozygosity Mapping Consortium for Autism, Nieto M, Walsh CA. Cell. 2016 Oct 6;167(2):341-354.
Links:
Christopher Walsh Laboratory (Boston Children’s Hospital and Harvard Medical School)
The Paul G. Allen Foundation Frontiers Group (Seattle)
NIH Support: National Institute of Neurological Disorders and Stroke; National Institute of Mental Health; National Institute of General Medical Sciences; National Cancer Institute
New Evidence Suggests Aging Brains Continue to Make New Neurons
Posted on by Dr. Francis Collins
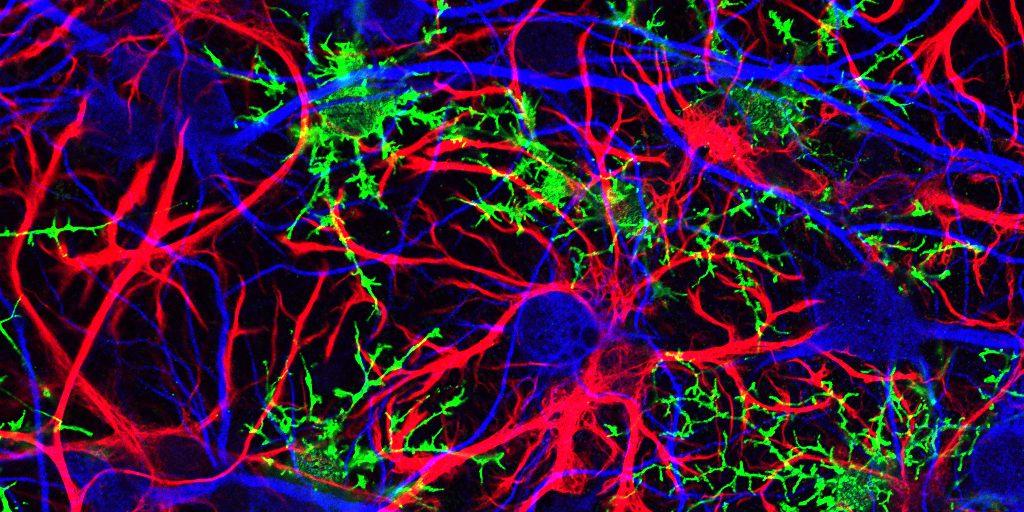
Caption: Mammalian hippocampal tissue. Immunofluorescence microscopy showing neurons (blue) interacting with neural astrocytes (red) and oligodendrocytes (green).
Credit: Jonathan Cohen, Fields Lab, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH
There’s been considerable debate about whether the human brain has the capacity to make new neurons into adulthood. Now, a recently published study offers some compelling new evidence that’s the case. In fact, the latest findings suggest that a healthy person in his or her seventies may have about as many young neurons in a portion of the brain essential for learning and memory as a teenager does.
As reported in the journal Cell Stem Cell, researchers examined the brains of healthy people, aged 14 to 79, and found similar numbers of young neurons throughout adulthood [1]. Those young neurons persisted in older brains that showed other signs of decline, including a reduced ability to produce new blood vessels and form new neural connections. The researchers also found a smaller reserve of quiescent, or inactive, neural stem cells in a brain area known to support cognitive-emotional resilience, the ability to cope with and bounce back from stressful circumstances.
While more study is clearly needed, the findings suggest healthy elderly people may have more cognitive reserve than is commonly believed. However, the findings may also help to explain why even perfectly healthy older people often find it difficult to face new challenges, such as travel or even shopping at a different grocery store, that wouldn’t have fazed them earlier in life.
Treating Zika Infection: Repurposed Drugs Show Promise
Posted on by Dr. Francis Collins

Credit: National Center for Advancing Translational Sciences, NIH
In response to the health threat posed by the recent outbreak of Zika virus in Latin America and its recent spread to Puerto Rico and Florida, researchers have been working at a furious pace to learn more about the mosquito-borne virus. Considerable progress has been made in understanding how Zika might cause babies to be born with unusually small heads and other abnormalities and in developing vaccines that may guard against Zika infection.
Still, there remains an urgent need to find drugs that can be used to treat people already infected with the Zika virus. A team that includes scientists at NIH’s National Center for Advancing Translational Sciences (NCATS) now has some encouraging news on this front. By testing 6,000 FDA-approved drugs and experimental chemical compounds on Zika-infected human cells in the lab, they’ve shown that some existing drugs might be repurposed to fight Zika infection and prevent the virus from harming the developing brain [1]. While additional research is needed, the new findings suggest it may be possible to speed development and approval of new treatments for Zika infection.
If I Only Had a Brain? Tissue Chips Predict Neurotoxicity
Posted on by Dr. Francis Collins
Caption: 3D neural tissue chips contain neurons (green), glial cells (red), and nuclei (blue). To take this confocal micrograph, developing neural tissue was removed from a chip and placed on a glass-bottom Petri dish.
Credit: Michael Schwartz, Dept. of Bioengineering, University of Wisconsin-Madison
A lot of time, money, and effort are devoted to developing new drugs. Yet only one of every 10 drug candidates entering human clinical trials successfully goes on to receive approval from the Food and Drug Administration (FDA) [1]. Many would-be drugs fall by the wayside because they prove toxic to the brain, liver, kidneys, or other organs—toxicity that, unfortunately, isn’t always detected in preclinical studies using mice, rats, or other animal models. That explains why scientists are working so hard to devise technologies that can do a better job of predicting early on which chemical compounds will be safe in humans.
As an important step in this direction, NIH-funded researchers at the Morgridge Institute for Research and University of Wisconsin-Madison have produced neural tissue chips with many features of a developing human brain. Each cultured 3D “organoid”—which sits comfortably in the bottom of a pea-sized well on a standard laboratory plate—comes complete with its very own neurons, support cells, blood vessels, and immune cells! As described in Proceedings of the National Academy of Sciences [2], this new tool is poised to predict earlier, faster, and less expensively which new or untested compounds—be they drug candidates or even ingredients in cosmetics and pesticides—might harm the brain, particularly at the earliest stages of development.
