Science
Study Suggests Treatments that Unleash Immune Cells in the Brain Could Help Combat Alzheimer’s
Posted on by Dr. Monica M. Bertagnolli
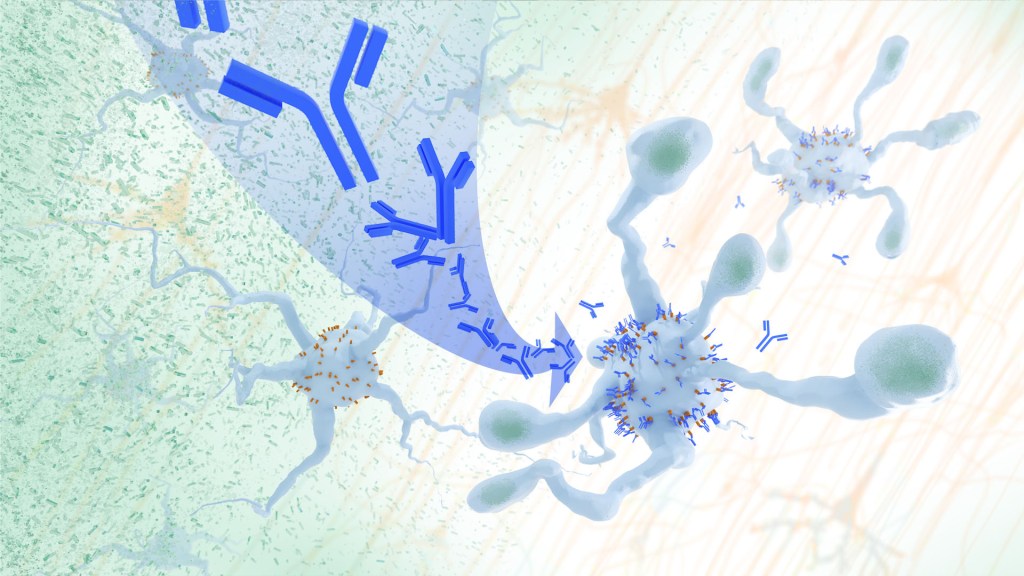
In Alzheimer’s disease, a buildup of sticky amyloid proteins in the brain clump together to form plaques, causing damage that gradually leads to worsening dementia symptoms. A promising way to change the course of this disease is with treatments that clear away damaging amyloid plaques or stop them from forming in the first place. In fact, the Food and Drug Administration recently approved the first drug for early Alzheimer’s that moderately slows cognitive decline by reducing amyloid plaques.1 Still, more progress is needed to combat this devastating disease that as many as 6.7 million Americans were living with in 2023.
Recent findings from a study in mice, supported in part by NIH and reported in Science Translational Medicine, offer another potential way to clear amyloid plaques in the brain. The key component of this strategy is using the brain’s built-in cleanup crew for amyloid plaques and other waste products: immune cells known as microglia that naturally help to limit the progression of Alzheimer’s. The findings suggest it may be possible to develop immunotherapies—treatments that use the body’s immune system to fight disease—to activate microglia in the brains of people with Alzheimer’s and clear amyloid plaques more effectively.2
In their report, the research team—including Marco Colonna, Washington University School of Medicine in St. Louis, and Jinchao Hou, now at Children’s Hospital of Zhejiang University School of Medicine in Zhejiang Province, China—wrote that microglia in the brain surround plaques to create a barrier that controls their spread. Microglia can also destroy amyloid plaques directly. But how microglia work in the brain depends on a fine-tuned balance of signals that activate or inhibit them. In people with Alzheimer’s, microglia don’t do their job well enough.
The researchers suspected this might have something to do with a protein called apolipoprotein E (APOE). This protein normally helps carry cholesterol and other fats in the bloodstream. But the gene encoding the protein is known for its role in influencing a person’s risk for developing Alzheimer’s, and in the Alzheimer’s brain, the protein is a key component of amyloid plaques. The protein can also inactivate microglia by binding to a receptor called LILRB4 found on the immune cells’ surfaces.
Earlier studies in mouse models of Alzheimer’s showed that the LILRB4 receptor is expressed at high levels in microglia when amyloid plaques build up. This suggested that treatments targeting this receptor on microglia might hold promise for treating Alzheimer’s. In the new study, the research team looked for evidence that an increase in LILRB4 receptors on microglia plays an important role in the brains of people with Alzheimer’s.
To do this, the researchers first studied brain tissue samples from people who died with this disease and discovered unusually high amounts of the LILRB4 receptor on the surfaces of microglia, similar to what had been seen in the mouse models. This could help explain why microglia struggle to control amyloid plaques in the Alzheimer’s brain.
Next, the researchers conducted studies of mouse brains with accumulating amyloid plaques that express the LILRB4 receptor to see if an antibody targeting the receptor could lower amyloid levels by boosting activity of immune microglia. Their findings suggest that the antibody treatment blocked the interaction between APOE proteins and LILRB4 receptors and enabled microglia to clear amyloid plaques. Intriguingly, the team’s additional studies found that this clearing process also changed the animals’ behavior, making them less likely to take risks. That’s important because people with Alzheimer’s may engage in risky behaviors as they lack memories of earlier experiences that they could use to make decisions.
There’s plenty more to learn. For instance, the researchers don’t know yet whether this approach will affect the tau protein, which forms damaging tangles inside neurons in the Alzheimer’s brain. They also want to investigate whether this strategy of clearing amyloid plaques might come with other health risks.
But overall, these findings add to evidence that immunotherapies of this kind could be a promising way to treat Alzheimer’s. This strategy may also have implications for treating other neurodegenerative conditions characterized by toxic debris in the brain, such as Parkinson’s disease, amyotrophic lateral sclerosis (ALS), and Huntington’s disease. The hope is that this kind of research will ultimately lead to more effective treatments for Alzheimer’s and other conditions affecting the brain.
References:
[1] FDA Converts Novel Alzheimer’s Disease Treatment to Traditional Approval. U.S. Food and Drug Administration (2023).
[2] Hou J, et al. Antibody-mediated targeting of human microglial leukocyte Ig-like receptor B4 attenuates amyloid pathology in a mouse model. Science Translational Medicine. DOI: 10.1126/scitranslmed.adj9052 (2024).
NIH Support: National Institute of General Medical Sciences, National Institute on Aging
Diagnosis and Treatment of Mental Health Conditions During and After Pregnancy is On the Rise, But Disparities Still Exist
Posted on by Dr. Monica M. Bertagnolli

Pregnancy and childbirth are often thought of as joyful times. Yet, we know that mental health conditions including perinatal depression, anxiety, and post-traumatic stress disorder (PTSD) are common complications during and after pregnancy, and this is contributing to a maternal health crisis in this country.
Now, a trio of NIH-supported studies reported in the journal Health Affairs show that diagnosis and treatment of mental health conditions such as anxiety, depression, and PTSD during pregnancy and in the first year after giving birth rose significantly in Americans with private health insurance from 2008 to 2020.1,2,3 While these are encouraging signs of increasing mental health awareness and service use, these studies also showed that this increase hasn’t happened equally across all demographic groups and states, making it clear there’s more work to do to ensure that people from all walks of life have access to the care they need, regardless of their race, ethnicity, geographic location, financial status, or other factors.
The findings come from a research team including Kara Zivin and Stephanie Hall and their colleagues at the University of Michigan, Ann Arbor. They recognized a worrying crisis in maternal mental illness in the U.S., with serious health risks and many potential long-term negative impacts for new parents and their infants. While earlier studies had looked at the prevalence of mental health conditions in the perinatal period, Zivin and Hall wanted to explore nationwide trends in the diagnosis of PTSD and what they refer to as perinatal mood and anxiety disorders (PMAD) among people giving birth between ages 15 and 44.
In the first study, using a database of administrative medical claims representing insured people across the U.S., the researchers examined PMAD diagnoses (including depressive and anxiety disorders) among those with private health insurance and found that the diagnosis rate had increased by more than 93% over the 12-year period under study. The rates also showed a sharp uptick after 2015, after the Affordable Care Act (ACA) went into effect in 2014. By 2020, 28% of those who were pregnant or in their postpartum period received a diagnosis of PMAD.
The researchers found that rate of suicidality (that is, suicidal ideation or diagnoses of self-harm) among people during pregnancy and just after childbirth more than doubled over the same period, although that rate dipped among those who had received a PMAD diagnosis. The rate at which individuals who were pregnant or in their postpartum period received any form of talk therapy covered by their private insurance increased by 16% from 2008 to 2020.
The second report focused specifically on PTSD diagnoses among privately insured people over the same period. The data show a near quadrupling of PTSD diagnoses in the months surrounding childbirth, with diagnoses in nearly 2% of privately insured people by 2020. Most of the increase in PTSD diagnoses occurred in people who had already been diagnosed with PMAD.
In the third study, the researchers wanted to learn if there were increases in antidepressant prescriptions for those diagnosed with PMAD, especially after new guidelines from several professional organizations were issued in 2015 and 2016. The findings show a decrease of 3% per year from 2008 to 2016, followed by a 32% increase in 2017, with prescription rates continuing to climb each year through 2020. By 2020, slightly less than half of those diagnosed with PMAD received a prescription for an antidepressant, suggesting that the clinical recommendations made a difference in clinical practice. However, there are signs of racial disparities, with White people with PMAD diagnoses receiving antidepressants more often than people in other racial groups.
In fact, all three studies show differences between people in different age, race, ethnicity, and geographic groups. For example, White people were more likely to be diagnosed with PTSD during the perinatal period, followed by Black people. By comparison, people of unknown race, as well as people who identified as Hispanic and Asian, were diagnosed less often. At the same time, the largest increase in PMAD diagnoses among races and ethnicities was among Black people, increasing from 14% of deliveries in 2008 to 22% in 2020.
Looking at age groups, the initial prevalence of PMAD diagnoses was highest among people aged 40 to 44, yet the youngest people (aged 15 to 24) experienced the largest increase in diagnoses. The youngest age group also had the largest increases in antidepressant prescriptions, and those aged 15 to 26 were more likely to be diagnosed with PTSD than those in older age groups. The first study also showed wide variation between states in the rate of PMAD diagnoses before and after implementation of the ACA.
The findings suggest there have been improvements in doctors’ recognition and treatment of PMAD and PTSD in the months surrounding childbirth. Increases in health insurance coverage and laws requiring equal treatment of mental health conditions may also be playing a role, along with greater social awareness and acceptance of mental health conditions generally. The researchers noted, however, that the findings don’t represent people with government-funded health insurance or those who lack health insurance completely.
It will be important to learn in future studies more about those who may still not be receiving the mental health care they need. The researchers report plans to look deeper into changes that have taken place at the state level and the impact of the pandemic and the rise of telehealth since 2020. Other recent NIH-supported research suggests that relatively straightforward interventions to reduce postpartum anxiety and depression can be remarkably effective. The key step will be not only identifying interventions that work, but also figuring out how to deliver effective treatments to the people who need them.
References:
[1] Zivin K, et al. Perinatal Mood and Anxiety Disorders Rose Among Privately Insured People, 2008-20. Health Affairs. DOI: 10.1377/hlthaff.2023.01437 (2024).
[2] Hall SV, et al. Perinatal Posttraumatic Stress Disorder Diagnoses Among Commercially Insured People Increased, 2008-20. Health Affairs. DOI: 10.1377/hlthaff.2023.01447 (2024).
[3] Hall SV, et al. Antidepressant Prescriptions Increased for Privately Insured People With Perinatal Mood And Anxiety Disorder, 2008-20. Health Affairs. DOI: 10.1377/hlthaff.2023.01448 (2024).
NIH Support: National Institute of Mental Health, National Institute on Minority Health and Health Disparities
Microbe Normally Found in the Mouth May Drive Progression of Colorectal Cancer
Posted on by Dr. Monica M. Bertagnolli

Colorectal cancer is a leading cause of death from cancer in the United States. We know that risk of colorectal cancer goes up with age, certain coexisting health conditions, family history, smoking, alcohol use, and other factors. Researchers are also trying to learn more about what leads colorectal cancer to grow and spread. Now, findings from a new study supported in part by NIH add to evidence that colorectal tumor growth may be driven by a surprising bad actor: a microbe that’s normally found in the mouth.1
The findings, reported in Nature, suggest that a subtype of the bacterium Fusobacterium nucleatum has distinct genetic properties that may allow it to withstand acidic conditions in the stomach, infect colorectal tumors, and potentially drive their growth, which may lead to poorer patient outcomes. The discoveries suggest that the microbe could eventually be used as a target for detecting and treating colorectal cancer.
The study was conducted by a team led by Susan Bullman and Christopher D. Johnston at the Fred Hutchinson Cancer Center in Seattle. In 2022, the team published findings from a pair of studies implicating Fusobacterium nucleatum in the progression and spread of colorectal cancer.2,3 Their findings weren’t the first to suggest a link between the microbe and colorectal cancer. But their work offered important evidence that the microbe might alter colorectal tumors in ways that made them more likely to grow and spread. They also found that the microbe may affect the way colorectal cancer responds to or resists chemotherapy treatment.
Follow-up studies suggested there might be more to the story, pointing to the possibility that certain strains of the bacterium might differ from others in important ways. The findings suggested that there may be a more specific subtype, not yet defined, that was responsible for driving colorectal cancer growth.
To look deeper into this in the new study, Bullman and Johnston, with first author Martha Zepeda Rivera, analyzed a collection of 55 strains of the microbe taken from human colorectal cancer samples. They also compared these at the genetic level to another 80 strains of the microbe taken from the mouths of people who didn’t have cancer.
Their studies uncovered 483 genetic factors that turned up more often in Fusobacterium nucleatum from colorectal tumors. Those strains mainly belonged to a subspecies called Fusobacterium nucleatum animalis (Fna). More detailed study led to another surprise. The Fna included two genetically distinct groups or “clades” that had never been described, which the researchers called Fna C1 and C2. It turned out that only Fna C2 occurs at high levels in colorectal tumors.
The researchers found that this specific subtype within colorectal tumors carries 195 genetic factors that may allow it to grow more rapidly, withstand the acidic environment in the stomach, and take up residence in the gastrointestinal tract, where it can drive colorectal cancer growth. When the researchers infected a mouse model of colitis, a condition involving inflamed intestines that is a risk factor for colorectal cancer, they found that Fna C2 caused the development of more tumors compared to those infected with Fna C1.
Studies of tumors from 116 patients with colorectal cancer also showed more Fna C2. It was elevated in about 50% of cases. In fact, only this strain turned up more often in cancer compared to healthy tissue nearby. Stool samples of 627 people with colorectal cancer and 619 healthy people also showed more of this specific microbial strain in association with cancer.
This discovery is important because it suggests it’s only the Fna C2 subtype that’s associated with driving colorectal tumor growth, meaning it could help in the development of new methods for colorectal cancer screening and treatment. The researchers suggest it may one day even be possible to develop microbial-based therapies using modified versions of the bacterial strain to deliver treatments straight into tumors.
In addition, while the microbe is normally found in healthy mouths, it’s also enriched in periodontal (gum) disease, dental infections, and oral cancers.4 It will be interesting to learn more in future studies about the connections between various Fusobacterium nucleatum subtypes, oral health, and other health conditions throughout the body, including colorectal cancer.
References:
[1] Zepeda-Rivera M, et al. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature. DOI: 10.1038/s41586-024-07182-w (2024).
[2] LaCourse KD, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. DOI: 10.1016/j.celrep.2022.111625 (2022).
[3] Galeano Niño JL, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. DOI: 10.1038/s41586-022-05435-0 (2022).
[4] Chen Y, et al. More Than Just a Periodontal Pathogen –the Research Progress on Fusobacterium nucleatum. Front Cell Infect Microbiol. DOI: 10.3389/fcimb.2022.815318 (2022).
NIH Support: National Institute of Dental and Craniofacial Research, National Cancer Institute
Visiting West Virginia University and Sharing Vision for Enhancing the Nation’s Health
Posted on by Dr. Monica M. Bertagnolli

NIH Director Monica Bertagnolli with Senator Shelley Moore Capito (left), and Dr. Ming Lei, Vice Dean of Research at the WVU School of Medicine (right), during a visit to West Virginia University Health Sciences Campus on March 27, 2024. Credit: (all photos) The Office of U.S. Senator Shelley Moore Capito (R-W.Va.).
In March, I had the pleasure of joining Senator Shelley Moore Capito in my first ever visit to West Virginia University Health Sciences Campus in Morgantown, where I got to witness NIH-supported research in action. On our tour, we visited a number of labs and scientific centers, learning about work being done in important areas of research including stroke, cancer, sleep trials, vaccine development, diabetes, cardiovascular heath, and more. I also gave a presentation in which I shared my vision, discussing my guiding principles and priorities as NIH Director, and I spoke about some of the biggest challenges in American health care and what NIH might do to address them. We were joined by Senator Joe Manchin and many distinguished WVU leaders and researchers, and I’m so grateful for the time and warm welcome they provided. To see such wide-ranging and vital research being done was inspiring, and I look forward to similar visits in the future.
The tour included a visit to WVU Cancer Institute’s mobile lung cancer screening unit, LUCAS, which travels the state, bringing lung cancer screening to people in rural areas.

Fear Switch in the Brain May Point to Target for Treating Anxiety Disorders Including PTSD
Posted on by Dr. Monica M. Bertagnolli

There’s a good reason you feel fear creep in when you’re walking alone at night in an unfamiliar place or hear a loud and unexpected noise ring out. In those moments, your brain triggers other parts of your nervous system to set a stress response in motion throughout your body. It’s that fear-driven survival response that keeps you alert, ready to fight or flee if the need arises. But when acute anxiety or traumatic events lead to fear that becomes generalized—occurring often and in situations that aren’t threatening—this can lead to debilitating anxiety disorders, including post-traumatic stress disorder (PTSD).
Just what happens in the brain’s circuitry to turn a healthy fear response into one that’s harmful hasn’t been well understood. Now, research findings by a team led by Nicholas Spitzer and Hui-Quan Li at the University of California San Diego and reported in the journal Science have pinpointed changes in the biochemistry of the brain and neural circuitry that lead to generalized fear.1 The intriguing findings, from research supported in part by NIH, raise the possibility that it might be possible to prevent or reverse this process with treatments targeting this fear “switch.”
To investigate generalized fear in the brain, the researchers first studied mice in the lab, looking at parts of the brain known to be linked to panic-like fear responses, including an area of the brainstem known as the dorsal raphe. They found that, in the mouse brain, acute stress led to a switch in the chemical messengers, or neurotransmitters, in some neurons within this portion of the mouse brain. Specifically, the chemical signals in the neurons flipped from producing excitatory glutamate neurotransmitters to inhibitory GABA neurotransmitters, and this led to a generalized fear response. They also found that the neurons that had undergone this switch are connected to brain regions that are known to play a role in fear responses including the amygdala and lateral hypothalamus. Interestingly, the researchers also showed they could avert generalized fear responses by preventing the production of GABA in the mouse brain.
To further support their research, the study team then examined postmortem brains of people who had PTSD and confirmed a similar switch in neurotransmitters to what happened in the mice. Next, they wanted to find out if they could block the switch by treating mice with the commonly used antidepressant fluoxetine. They found that when mice were treated with fluoxetine in their drinking water promptly after a stressful event, the neurotransmitter switch and subsequent generalized fear were prevented.
The researchers made even more findings about the timing of the switch that could lead to better treatments. They found that in mice, the switch to generalized fear persisted for four weeks after an acutely stressful event—a period that for the mice may be the equivalent of three years in people. This suggests that treatments may prevent generalized fear and the development of anxiety disorders when given before the brain undergoes a neurotransmitter switch. The findings may also explain why treatment doesn’t seem to be as effective in people who are initially treated for PTSD after having it for a long time.
Going forward, the researchers want to explore targeted approaches to reversing this fear switch after it has taken place. The hope is to discover new ways to rid the brain of generalized fear responses and help treat anxiety disorders including PTSD, a condition which will affect more than six in every 100 people at some point in their lives.2
References:
[1] Li HQ, et al. Generalized fear after acute stress is caused by change in neuronal cotransmitter identity. Science. DOI: 10.1126/science.adj5996 (2024).
[2] Post-Traumatic Stress Disorder (PTSD). National Institute of Mental Health.
NIH Support: National Institute of Neurological Disorders and Stroke
Immune Checkpoint Discovery Has Implications for Treating Cancer and Autoimmune Diseases
Posted on by Dr. Monica M. Bertagnolli
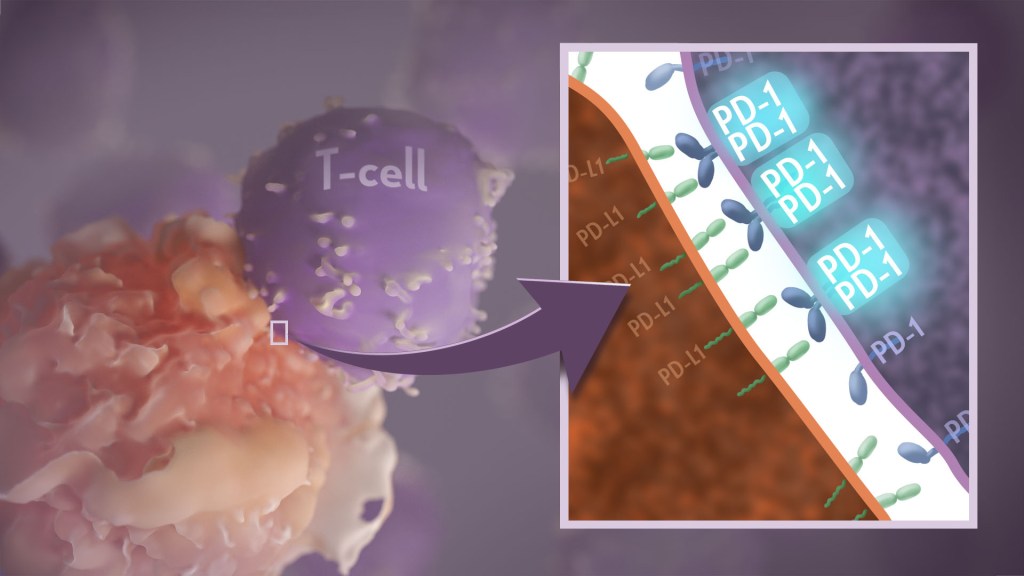
Your immune system should ideally recognize and attack infectious invaders and cancerous cells. But the system requires safety mechanisms, or brakes, to keep it from damaging healthy cells. To do this, T cells—the immune system’s most powerful attackers—rely on immune “checkpoints” to turn immune activation down when they receive the right signal. While these interactions have been well studied, a research team supported in part by NIH has made an unexpected discovery into how a key immune checkpoint works, with potentially important implications for therapies designed to boost or dampen immune activity to treat cancer and autoimmune diseases.1
The checkpoint in question is a protein called programmed cell death-1 (PD-1). Here’s how it works: PD-1 is a receptor on the surface of T cells, where it latches onto certain proteins, known as PD-L1 and PD-L2, on the surface of other cells in the body. When this interaction occurs, a signal is sent to the T cells that stops them from attacking these other cells.
Cancer cells often take advantage of this braking system, producing copious amounts of PD-L1 on their surface, allowing them to hide from T cells. An effective class of immunotherapy drugs used to treat many cancers works by blocking the interaction between PD-1 and PD-L1, to effectively release the brakes on the immune system to allow the T cells to unleash an assault on cancer cells. Researchers have also developed potential treatments for autoimmune diseases that take the opposite tact: stimulating PD-1 interaction to keep T cells inactive. These PD-1 “agonists” have shown promise in clinical trials as treatments for certain autoimmune diseases.
However, to fulfill the promise of these approaches for treating cancer and autoimmune diseases, a better understanding of precisely how PD-1 works to suppress T cell activity is still needed. The thinking has long been that individual PD-1 receptors act alone. But, as reported in Science Immunology, it turns out that this may not usually be the case. A team led by Jun Wang and Xiangpeng Kong of New York University Langone Health’s Perlmutter Cancer Center, with Elliot Philips of NYU and Michael Dustin of the University of Oxford, U.K., used sophisticated techniques to look for evidence of what happens when PD-1 proteins work together in pairs.
They found that PD-1’s tendency to link, or not link, with a second PD-1 protein to form what’s known as a “dimer” depends on interactions with portions of the protein that cross the immune cell membrane. They also found that, when PD-1 receptors pair up, they do a better job of squashing immune responses. The findings also showed that a single change in the amino acid structure in the portion of PD-1 that crosses the cell membrane can strengthen or weaken immune responses.
One reason why these fundamental discoveries are exciting is they suggest that interfering with PD-1’s ability to form dimers might make immunotherapy treatments for cancer more effective. In addition, treatments that strengthen interactions between paired PD-1 receptors might aid in the design of promising new drug classes that are intended to tamp down inflammation seen in people with some autoimmune diseases, including rheumatoid arthritis, lupus, and type 1 diabetes. The research team now plans to conduct further investigations of PD-1 blockers and agonists to explore whether these findings could eventually lead to more effective treatments for both cancer and autoimmune diseases.
Reference:
[1] Philips EA, et al. Transmembrane domain-driven PD-1 dimers mediate T cell inhibition. Science Immunology. DOI: 10.1126/sciimmunol.ade6256 (2024).
NIH Support: National Institute of Allergy and Infectious Diseases, National Cancer Institute, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institute of General Medical Sciences
Welcoming Internet Pioneer Vint Cerf for Rall Cultural Lecture on AI in Biomedical Research
Posted on by Dr. Monica M. Bertagnolli

Last week it was my pleasure to welcome to NIH Vinton “Vint” Cerf, a pioneer of the digital world widely known as one of the “fathers of the internet,” to speak at our annual J. Edward Rall Cultural Lecture. We had a lively fireside chat focusing on “The Promises and Perils of AI in Biomedical Research and Health Care Delivery” that included discussions on topics like improving data collection, ensuring broad representativeness of the data being used to train AI and machine learning, expanding clinical research, and offering advice for scientists just starting out. It was an insightful, eye-opening conversation on a topic close to my heart. As NIH Director, I very much hope to deliver evidence-based health care to all people, and I’m excited about how AI and machine learning can help us advance toward that goal.
Dr. Cerf, who is vice president and Chief Internet Evangelist for Google, has too many awards to list, but just a few include the U.S. National Medal of Technology, the Turing Award, and the Presidential Medal of Freedom. He serves as an advisor to several government agencies, including the National Science Foundation, NASA, and the Departments of Defense, Energy, and Commerce.

The NIH Rall Cultural Lecture is held in honor of Dr. J. Edward Rall, who helped define the modern intramural research program at NIH. In the 1950s, he helped establish a stable, academic-minded, and culturally rich community within our rapidly expanding government agency. In 1984, he recommended that NIH add a cultural lecture to its Director’s Lecture series to enrich our scientific community.
As I said at the event, I’m grateful to Dr. Cerf for his visit and this conversation, and I can’t think of a more spectacular Rall Cultural Lecture than what he provided for all of us. You can watch the full talk here.

Study Suggests During Sleep, Neural Process Helps Clear the Brain of Damaging Waste
Posted on by Dr. Monica M. Bertagnolli
We’ve long known that sleep is a restorative process necessary for good health. Research has also shown that the accumulation of waste products in the brain is a leading cause of numerous neurological disorders, including Alzheimer’s and Parkinson’s diseases. What hasn’t been clear is how the healthy brain “self-cleans,” or flushes out that detrimental waste.
But a new study by a research team supported in part by NIH suggests that a neural process that happens while we sleep helps cleanse the brain, leading us to wake up feeling rested and restored. Better understanding this process could one day lead to methods that help people function well on less sleep. It could also help researchers find potential ways to delay or prevent neurological diseases related to accumulated waste products in the brain.
The findings, reported in Nature, show that, during sleep, neural networks in the brain act like an array of miniature pumps, producing large and rhythmic waves through synchronous bursts of activity that propel fluids through brain tissue. Much like the process of washing dishes, where you use a rhythmic motion of varying speeds and intensity to clear off debris, this process that takes place during sleep clears accumulated metabolic waste products out.
The research team, led by Jonathan Kipnis and Li-Feng Jiang-Xie at Washington University School of Medicine in St. Louis, wanted to better understand how the brain manages its waste. This is not an easy task, given that the human brain’s billions of neurons inevitably produce plenty of junk during cognitive processes that allow us to think, feel, move, and solve problems. Those waste products also build in a complex environment, including a packed maze of interconnected neurons, blood vessels, and interstitial spaces, surrounded by a protective blood-brain barrier that limits movement of substances in or out.
So, how does the brain move fluid through those tight spaces with the force required to get waste out? Earlier research suggested that neural activity during sleep might play an important role in those waste-clearing dynamics. But previous studies hadn’t pinned down the way this works.
To learn more in the new study, the researchers recorded brain activity in mice. They also used an ultrathin silicon probe to measure fluid dynamics in the brain’s interstitial spaces. In awake mice, they saw irregular neural activity and only minor fluctuations in the interstitial spaces. But when the animals were resting under anesthesia, the researchers saw a big change. Brain recordings showed strongly enhanced neural activity, with two distinct but tightly coupled rhythms. The research team realized that the structured wave patterns could generate strong energy that could move small molecules and peptides, or waste products, through the tight spaces within brain tissue.
To make sure that the fluid dynamics were really driven by neurons, the researchers used tools that allowed them to turn neural activity off in some areas. Those experiments showed that, when neurons stopped firing, the waves also stopped. They went on to show similar dynamics during natural sleep in the animals and confirmed that disrupting these neuron-driven fluid dynamics impaired the brain’s ability to clear out waste.
These findings highlight the importance of this cleansing process during sleep for brain health. The researchers now want to better understand how specific patterns and variations in those brain waves lead to changes in fluid movement and waste clearance. This could help researchers eventually find ways to speed up the removal of damaging waste, potentially preventing or delaying certain neurological diseases and allowing people to need less sleep.
Reference:
[1] Jiang-Xie LF, et al. Neuronal dynamics direct cerebrospinal fluid perfusion and brain clearance. Nature. DOI: 10.1038/s41586-024-07108-6 (2024).
NIH Support: National Center for Complementary and Integrative Health
Healing Switch Links Acute Kidney Injury to Fibrosis, Suggesting Way to Protect Kidney Function
Posted on by Dr. Monica M. Bertagnolli
Healthy kidneys—part of the urinary tract—remove waste and help balance chemicals and fluids in the body. However, our kidneys have a limited ability to regenerate healthy tissue after sustaining injuries from conditions such as diabetes or high blood pressure. Injured kidneys are often left with a mix of healthy and scarred tissue, or fibrosis, which over time can compromise their function and lead to chronic kidney disease or complete kidney failure. More than one in seven adults in the U.S. are estimated to have chronic kidney disease, according to the Centers for Disease Control and Prevention, most without knowing it.
Now, a team of researchers led by Sanjeev Kumar at Cedars-Sinai Medical Center, Los Angeles, has identified a key molecular “switch” that determines whether injured kidney tissue will heal or develop those damaging scars.1 Their findings, reported in the journal Science, could lead to new and less invasive ways to detect fibrosis in the kidneys. The research could also point toward a targeted therapeutic approach that might prevent or reverse scarring to protect kidney function.
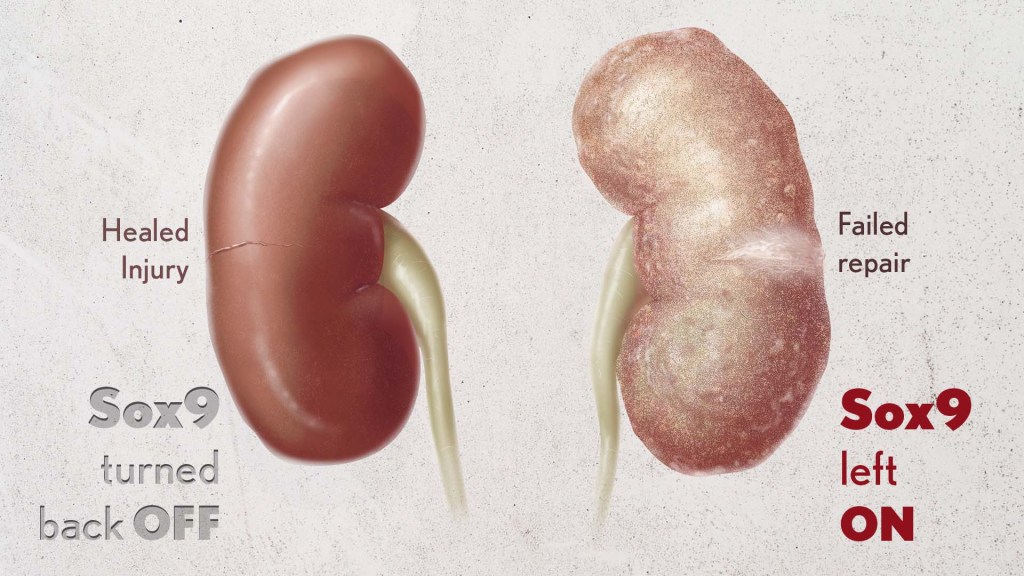
In earlier studies, the research team found that a protein called Sox9 plays an important role in switching on the repair response in kidneys after acute injury.2 In some cases, the researchers noticed that Sox9 remained active for a prolonged period of a month or more. They suspected this might be a sign of unresolved injury and repair.
By conducting studies using animal models of kidney damage, the researchers found that cells that turned Sox9 on and then back off healed without fibrosis. However, cells that failed to regenerate healthy kidney cells kept Sox9 on indefinitely, which in turn led to the production of fibrosis and scarring.
According to Kumar, Sox9 appears to act like a sensor, switching on after injury. Once restored to health, Sox9 switches back off. When healing doesn’t proceed optimally, Sox9 stays on, leading to scarring. Importantly, the researchers also found they could encourage kidneys to recover by forcing Sox9 to turn off a week after an injury, suggesting it may be a promising drug target.
The researchers also looked for evidence of this process in human patients who have received kidney transplants. They could see that, when transplanted kidneys took longer to start working, Sox9 was switched on. Those whose kidneys continued to produce Sox9 also had lower kidney function and more scarring compared to those who didn’t.
The findings suggest that the dynamics observed in animal studies may be clinically relevant in people, and that treatments targeting Sox9 might promote kidneys to heal instead of scarring. The researchers say they hope that similar studies in the future will lead to greater understanding of healing and fibrosis in other organs—including the heart, lungs, and liver—with potentially important clinical implications.
References:
[1] Aggarwal S, et al. SOX9 switch links regeneration to fibrosis at the single-cell level in mammalian kidneys. Science. DOI: 10.1126/science.add6371 (2024).
[2] Kumar S, et al. Sox9 Activation Highlights a Cellular Pathway of Renal Repair in the Acutely Injured Mammalian Kidney. Cell Reports. DOI: 10.1016/j.celrep.2015.07.034 (2015).
NIH Support: National Institute of Diabetes and Digestive and Kidney Diseases
Aided by AI, Study Uncovers Hidden Sex Differences in Dynamic Brain Function
Posted on by Dr. Monica M. Bertagnolli

We’re living in an especially promising time for biomedical discovery and advances in the delivery of data-driven health care for everyone. A key part of this is the tremendous progress made in applying artificial intelligence to study human health and ultimately improve clinical care in many important and sometimes surprising ways.1 One new example of this comes from a fascinating study, supported in part by NIH, that uses AI approaches to reveal meaningful sex differences in the way the brain works.
As reported in the Proceedings of the National Academy of Sciences, researchers led by Vinod Menon at Stanford Medicine, Stanford, CA, have built an AI model that can—nine times out of ten—tell whether the brain in question belongs to a female or male based on scans of brain activity alone.2 These findings not only help resolve long-term debates about whether reliable differences between sexes exist in the human brain, but they’re also a step toward improving our understanding of why some psychiatric and neurological disorders affect women and men differently.
The prevalence of certain psychiatric and neurological disorders in men and women can vary significantly, leading researchers to suspect that sex differences in brain function likely exist. For example, studies have found that females are more likely to experience depression, anxiety, and eating disorders, while autism, Attention-Deficit/Hyperactivity Disorder, and schizophrenia are seen more often in males. But earlier research to understand sex differences in the brain have focused mainly on anatomical and structural studies of brain regions and their connections. Much less is known about how those structural differences translate into differences in brain activity and function.
To help fill those gaps in the new study, Menon’s team took advantage of vast quantities of brain activity data from MRI scans from the NIH-supported Human Connectome Project. The data was captured from hundreds of healthy young adults with the goal of studying the brain and how it changes with growth, aging, and disease. To use this data to explore sex differences in brain function, the researchers developed what’s known as a deep neural network model in which a computer “learned” how to recognize patterns in brain activity data that could distinguish a male from a female brain.
This approach doesn’t rely on any preconceived notions about what features might be important. A computer is simply shown many examples of brain activity belonging to males and females and, over time, can begin to pick up on otherwise hidden differences that are useful for making such classifications accurately. One of the things that made this work different from earlier attempts was it relied on dynamic scans of brain activity, which capture the interplay among brain regions.
After analyzing about 1,500 brain scans, a computer could usually (although not always) tell whether a scan came from a male or female brain. The findings also showed the model worked reliably well in different datasets and in brain scans for people in different places in the U.S. and Europe. Overall, the findings confirm that reliable sex differences in brain activity do exist.
Where did the model find those differences? To get an idea, the researchers turned to an approach called explainable AI, which allowed them to dig deeper into the specific features and brain areas their model was using to pick up on sex differences. It turned out that one set of areas the model was relying on to distinguish between male and female brains is what’s known as the default mode network. This area is responsible for processing self-referential information and constructing a coherent sense of the self and activates especially when people let their minds wander.3 Other important areas included the striatum and limbic network, which are involved in learning and how we respond to rewards, respectively.
Many questions remain, including whether such differences arise primarily due to inherent biological differences between the sexes or what role societal circumstances play. But the researchers say that the discovery already shows that sex differences in brain organization and function may play important and overlooked roles in mental health and neuropsychiatric disorders. Their AI model can now also be applied to begin to explain other kinds of brain differences, including those that may affect learning or social behavior. It’s an exciting example of AI-driven progress and good news for understanding variations in human brain functions and their implications for our health.
References:
[1] Bertagnolli, MM. Advancing health through artificial intelligence/machine learning: The critical importance of multidisciplinary collaboration. PNAS Nexus. DOI: 10.1093/pnasnexus/pgad356 (2023).
[2] Ryali S, et al. Deep learning models reveal replicable, generalizable, and behaviorally relevant sex differences in human functional brain organization. Proc Natl Acad Sci. DOI: 10.1073/pnas.2310012121 (2024).
[3] Menon, V. 20 years of the default mode network: A review and synthesis. Neuron.DOI: 10.1016/j.neuron.2023.04.023 (2023).
NIH Support: National Institute of Mental Health, National Institute of Biomedical Imaging and Bioengineering, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute on Aging
Next Page
