2019 March
Largest-Ever Alzheimer’s Gene Study Brings New Answers
Posted on by Dr. Francis Collins

Predicting whether someone will get Alzheimer’s disease (AD) late in life, and how to use that information for prevention, has been an intense focus of biomedical research. The goal of this work is to learn not only about the genes involved in AD, but how they work together and with other complex biological, environmental, and lifestyle factors to drive this devastating neurological disease.
It’s good news to be able to report that an international team of researchers, partly funded by NIH, has made more progress in explaining the genetic component of AD. Their analysis, involving data from more than 35,000 individuals with late-onset AD, has identified variants in five new genes that put people at greater risk of AD [1]. It also points to molecular pathways involved in AD as possible avenues for prevention, and offers further confirmation of 20 other genes that had been implicated previously in AD.
The results of this largest-ever genomic study of AD suggests key roles for genes involved in the processing of beta-amyloid peptides, which form plaques in the brain recognized as an important early indicator of AD. They also offer the first evidence for a genetic link to proteins that bind tau, the protein responsible for telltale tangles in the AD brain that track closely with a person’s cognitive decline.
The new findings are the latest from the International Genomics of Alzheimer’s Project (IGAP) consortium, led by a large, collaborative team including Brian Kunkle and Margaret Pericak-Vance, University of Miami Miller School of Medicine, Miami, FL. The effort, spanning four consortia focused on AD in the United States and Europe, was launched in 2011 with the aim of discovering and mapping all the genes that contribute to AD.
An earlier IGAP study including about 25,500 people with late-onset AD identified 20 common gene variants that influence a person’s risk for developing AD late in life [2]. While that was terrific progress to be sure, the analysis also showed that those gene variants could explain only a third of the genetic component of AD. It was clear more genes with ties to AD were yet to be found.
So, in the study reported in Nature Genetics, the researchers expanded the search. While so-called genome-wide association studies (GWAS) are generally useful in identifying gene variants that turn up often in association with particular diseases or other traits, the ones that arise more rarely require much larger sample sizes to find.
To increase their odds of finding additional variants, the researchers analyzed genomic data for more than 94,000 individuals, including more than 35,000 with a diagnosis of late-onset AD and another 60,000 older people without AD. Their search led them to variants in five additional genes, named IQCK, ACE, ADAM10, ADAMTS1, and WWOX, associated with late-onset AD that hadn’t turned up in the previous study.
Further analysis of those genes supports a view of AD in which groups of genes work together to influence risk and disease progression. In addition to some genes influencing the processing of beta-amyloid peptides and accumulation of tau proteins, others appear to contribute to AD via certain aspects of the immune system and lipid metabolism.
Each of these newly discovered variants contributes only a small amount of increased risk, and therefore probably have limited value in predicting an average person’s risk of developing AD later in life. But they are invaluable when it comes to advancing our understanding of AD’s biological underpinnings and pointing the way to potentially new treatment approaches. For instance, these new data highlight intriguing similarities between early-onset and late-onset AD, suggesting that treatments developed for people with the early-onset form also might prove beneficial for people with the more common late-onset disease.
It’s worth noting that the new findings continue to suggest that the search is not yet over—many more as-yet undiscovered rare variants likely play a role in AD. The search for answers to AD and so many other complex health conditions—assisted through collaborative data sharing efforts such as this one—continues at an accelerating pace.
References:
[1] Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Kunkle BW, Grenier-Boley B, Sims R, Bis JC, et. al. Nat Genet. 2019 Mar;51(3):414-430.
[2] Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, et al. Nat Genet. 2013 Dec;45(12):1452-8.
Links:
Alzheimer’s Disease Genetics Fact Sheet (National Institute on Aging/NIH)
Genome-Wide Association Studies (NIH)
Margaret Pericak-Vance (University of Miami Health System, FL)
NIH Support: National Institute on Aging; National Heart, Lung, and Blood Institute; National Human Genome Research Institute; National Institute of Allergy and Infectious Diseases; Eunice Kennedy Shriver National Institute of Child Health and Human Development; National Institute of Diabetes and Digestive and Kidney Disease; National Institute of Neurological Disorders and Stroke
Finalists in Science Talent Search
Posted on by Dr. Francis Collins

Credit: Society for Science & the Public
A Warm Welcome to African Fellows
Posted on by Dr. Francis Collins

A Song for Steve
Posted on by Dr. Francis Collins

Can a Mind-Reading Computer Speak for Those Who Cannot?
Posted on by Dr. Francis Collins
Credit: Adapted from Nima Mesgarani, Columbia University’s Zuckerman Institute, New York
Computers have learned to do some amazing things, from beating the world’s ranking chess masters to providing the equivalent of feeling in prosthetic limbs. Now, as heard in this brief audio clip counting from zero to nine, an NIH-supported team has combined innovative speech synthesis technology and artificial intelligence to teach a computer to read a person’s thoughts and translate them into intelligible speech.
Turning brain waves into speech isn’t just fascinating science. It might also prove life changing for people who have lost the ability to speak from conditions such as amyotrophic lateral sclerosis (ALS) or a debilitating stroke.
When people speak or even think about talking, their brains fire off distinctive, but previously poorly decoded, patterns of neural activity. Nima Mesgarani and his team at Columbia University’s Zuckerman Institute, New York, wanted to learn how to decode this neural activity.
Mesgarani and his team started out with a vocoder, a voice synthesizer that produces sounds based on an analysis of speech. It’s the very same technology used by Amazon’s Alexa, Apple’s Siri, or other similar devices to listen and respond appropriately to everyday commands.
As reported in Scientific Reports, the first task was to train a vocoder to produce synthesized sounds in response to brain waves instead of speech [1]. To do it, Mesgarani teamed up with neurosurgeon Ashesh Mehta, Hofstra Northwell School of Medicine, Manhasset, NY, who frequently performs brain mapping in people with epilepsy to pinpoint the sources of seizures before performing surgery to remove them.
In five patients already undergoing brain mapping, the researchers monitored activity in the auditory cortex, where the brain processes sound. The patients listened to recordings of short stories read by four speakers. In the first test, eight different sentences were repeated multiple times. In the next test, participants heard four new speakers repeat numbers from zero to nine.
From these exercises, the researchers reconstructed the words that people heard from their brain activity alone. Then the researchers tried various methods to reproduce intelligible speech from the recorded brain activity. They found it worked best to combine the vocoder technology with a form of computer artificial intelligence known as deep learning.
Deep learning is inspired by how our own brain’s neural networks process information, learning to focus on some details but not others. In deep learning, computers look for patterns in data. As they begin to “see” complex relationships, some connections in the network are strengthened while others are weakened.
In this case, the researchers used the deep learning networks to interpret the sounds produced by the vocoder in response to the brain activity patterns. When the vocoder-produced sounds were processed and “cleaned up” by those neural networks, it made the reconstructed sounds easier for a listener to understand as recognizable words, though this first attempt still sounds pretty robotic.
The researchers will continue testing their system with more complicated words and sentences. They also want to run the same tests on brain activity, comparing what happens when a person speaks or just imagines speaking. They ultimately envision an implant, similar to those already worn by some patients with epilepsy, that will translate a person’s thoughts into spoken words. That might open up all sorts of awkward moments if some of those thoughts weren’t intended for transmission!
Along with recently highlighted new ways to catch irregular heartbeats and cervical cancers, it’s yet another remarkable example of the many ways in which computers and artificial intelligence promise to transform the future of medicine.
Reference:
[1] Towards reconstructing intelligible speech from the human auditory cortex. Akbari H, Khalighinejad B, Herrero JL, Mehta AD, Mesgarani N. Sci Rep. 2019 Jan 29;9(1):874.
Links:
Advances in Neuroprosthetic Learning and Control. Carmena JM. PLoS Biol. 2013;11(5):e1001561.
Nima Mesgarani (Columbia University, New York)
NIH Support: National Institute on Deafness and Other Communication Disorders; National Institute of Mental Health
Visiting University of Delaware
Posted on by Dr. Francis Collins

‘Nanoantennae’ Make Infrared Vision Possible
Posted on by Dr. Francis Collins
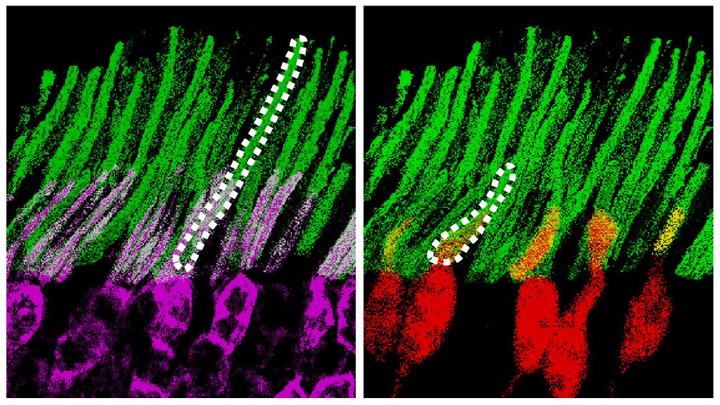
Credit: Ma et al. Cell, 2019
Infrared vision often brings to mind night-vision goggles that allow soldiers to see in the dark, like you might have seen in the movie Zero Dark Thirty. But those bulky goggles may not be needed one day to scope out enemy territory or just the usual things that go bump in the night. In a dramatic advance that brings together material science and the mammalian vision system, researchers have just shown that specialized lab-made nanoparticles applied to the retina, the thin tissue lining the back of the eye, can extend natural vision to see in infrared light.
The researchers showed in mouse studies that their specially crafted nanoparticles bind to the retina’s light-sensing cells, where they act like “nanoantennae” for the animals to see and recognize shapes in infrared—day or night—for at least 10 weeks. Even better, the mice maintained their normal vision the whole time and showed no adverse health effects. In fact, some of the mice are still alive and well in the lab, although their ability to see in infrared may have worn off.
When light enters the eyes of mice, humans, or any mammal, light-sensing cells in the retina absorb wavelengths within the range of visible light. (That’s roughly from 400 to 700 nanometers.) While visible light includes all the colors of the rainbow, it actually accounts for only a fraction of the full electromagnetic spectrum. Left out are the longer wavelengths of infrared light. That makes infrared light invisible to the naked eye.
In the study reported in the journal Cell, an international research team including Gang Han, University of Massachusetts Medical School, Worcester, wanted to find a way for mammalian light-sensing cells to absorb and respond to the longer wavelengths of infrared [1]. It turns out Han’s team had just the thing to do it.
His NIH-funded team was already working on the nanoparticles now under study for application in a field called optogenetics—the use of light to control living brain cells [2]. Optogenetics normally involves the stimulation of genetically modified brain cells with blue light. The trouble is that blue light doesn’t penetrate brain tissue well.
That’s where Han’s so-called upconversion nanoparticles (UCNPs) came in. They attempt to get around the normal limitations of optogenetic tools by incorporating certain rare earth metals. Those metals have a natural ability to absorb lower energy infrared light and convert it into higher energy visible light (hence the term upconversion).
But could those UCNPs also serve as miniature antennae in the eye, receiving infrared light and emitting readily detected visible light? To find out in mouse studies, the researchers injected a dilute solution containing UCNPs into the back of eye. Such sub-retinal injections are used routinely by ophthalmologists to treat people with various eye problems.
These UCNPs were modified with a protein that allowed them to stick to light-sensing cells. Because of the way that UCNPs absorb and emit wavelengths of light energy, they should to stick to the light-sensing cells and make otherwise invisible infrared light visible as green light.
Their hunch proved correct, as mice treated with the UCNP solution began seeing in infrared! How could the researchers tell? First, they shined infrared light into the eyes of the mice. Their pupils constricted in response just as they would with visible light. Then the treated mice aced a series of maneuvers in the dark that their untreated counterparts couldn’t manage. The treated animals also could rely on infrared signals to make out shapes.
The research is not only fascinating, but its findings may also have a wide range of intriguing applications. One could imagine taking advantage of the technology for use in hiding encrypted messages in infrared or enabling people to acquire a temporary, built-in ability to see in complete darkness.
With some tweaks and continued research to confirm the safety of these nanoparticles, the system might also find use in medicine. For instance, the nanoparticles could potentially improve vision in those who can’t see certain colors. While such infrared vision technologies will take time to become more widely available, it’s a great example of how one area of science can cross-fertilize another.
References:
[1] Mammalian Near-Infrared Image Vision through Injectable and Self-Powered Retinal Nanoantennae. Ma Y, Bao J, Zhang Y, Li Z, Zhou X, Wan C, Huang L, Zhao Y, Han G, Xue T. Cell. 2019 Feb 27. [Epub ahead of print]
[2] Near-Infrared-Light Activatable Nanoparticles for Deep-Tissue-Penetrating Wireless Optogenetics. Yu N, Huang L, Zhou Y, Xue T, Chen Z, Han G. Adv Healthc Mater. 2019 Jan 11:e1801132.
Links:
Diagram of the Eye (National Eye Institute/NIH)
Infrared Waves (NASA)
Visible Light (NASA)
Han Lab (University of Massachusetts, Worcester)
NIH Support: National Institute of Mental Health; National Institute of General Medical Sciences
Previous Page
