severe acute respiratory syndrome
‘Decoy’ Protein Works Against Multiple Coronavirus Variants in Early Study
Posted on by Lawrence Tabak, D.D.S., Ph.D.
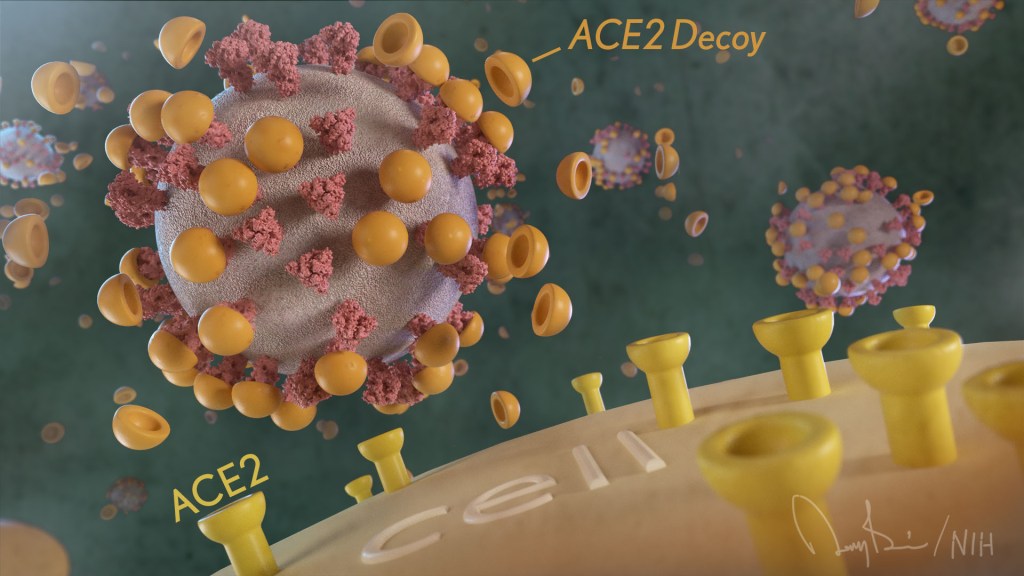
The NIH continues to support the development of some very innovative therapies to control SARS-CoV-2, the coronavirus that causes COVID-19. One innovative idea involves a molecular decoy to thwart the coronavirus.
How’s that? The decoy is a specially engineered protein particle that mimics the 3D structure of the ACE2 receptor, a protein on the surface of our cells that the virus’s spike proteins bind to as the first step in causing an infection.
The idea is when these ACE2 decoys are administered therapeutically, they will stick to the spike proteins that crown the coronavirus (see image above). With its spikes covered tightly in decoy, SARS-CoV-2 has a more-limited ability to attach to the real ACE2 and infect our cells.
Recently, the researchers published their initial results in the journal Nature Chemical Biology, and the early data look promising [1]. They found in mouse models of severe COVID-19 that intravenous infusion of an engineered ACE2 decoy prevented lung damage and death. Though more study is needed, the researchers say the decoy therapy could potentially be delivered directly to the lungs through an inhaler and used alone or in combination with other COVID-19 treatments.
The findings come from a research team at the University of Illinois Chicago team, led by Asrar Malik and Jalees Rehman, working in close collaboration with their colleagues at the University of Illinois Urbana-Champaign. The researchers had been intrigued by an earlier clinical trial testing the ACE2 decoy strategy [2]. However, in this earlier attempt, the clinical trial found no reduction in mortality. The ACE2 drug candidate, which is soluble and degrades in the body, also proved ineffective in neutralizing the virus.
Rather than give up on the idea, the UIC team decided to give it a try. They engineered a new soluble version of ACE2 that structurally might work better as a decoy than the original one. Their version of ACE2, which includes three changes in the protein’s amino acid building blocks, binds the SARS-CoV-2 spike protein much more tightly. In the lab, it also appeared to neutralize the virus as well as monoclonal antibodies used to treat COVID-19.
To put it to the test, they conducted studies in mice. Normal mice don’t get sick from SARS-CoV-2 because the viral spike can’t bind well to the mouse version of the ACE2 receptor. So, the researchers did their studies in a mouse that carries the human ACE2 and develops a severe acute respiratory syndrome somewhat similar to that seen in humans with severe COVID-19.
In their studies, using both the original viral isolate from Washington State and the Gamma variant (P.1) first detected in Brazil, they found that infected mice infused with their therapeutic ACE2 protein had much lower mortality and showed few signs of severe acute respiratory syndrome. While the protein worked against both versions of the virus, infection with the more aggressive Gamma variant required earlier treatment. The treated mice also regained their appetite and weight, suggesting that they were making a recovery.
Further studies showed that the decoy bound to spike proteins from every variant tested, including Alpha, Beta, Delta and Epsilon. (Omicron wasn’t yet available at the time of the study.) In fact, the decoy bound just as well, if not better, to new variants compared to the original virus.
The researchers will continue their preclinical work. If all goes well, they hope to move their ACE2 decoy into a clinical trial. What’s especially promising about this approach is it could be used in combination with treatments that work in other ways, such as by preventing virus that’s already infected cells from growing or limiting an excessive and damaging immune response to the infection.
Last week, more than 17,500 people in the United States were hospitalized with severe COVID-19. We’ve got to continue to do all we can to save lives, and it will take lots of innovative ideas, like this ACE2 decoy, to put us in a better position to beat this virus once and for all.
References:
[1] Engineered ACE2 decoy mitigates lung injury and death induced by SARS-CoV-2 variants.
Zhang L, Dutta S, Xiong S, Chan M, Chan KK, Fan TM, Bailey KL, Lindeblad M, Cooper LM, Rong L, Gugliuzza AF, Shukla D, Procko E, Rehman J, Malik AB. Nat Chem Biol. 2022 Jan 19.
[2] Recombinant human angiotensin-converting enzyme 2 (rhACE2) as a treatment for patients with COVID-19 (APN01-COVID-19). ClinicalTrials.gov.
Links:
COVID-19 Research (NIH)
Accelerating COVID-19 Therapeutic Interventions and Vaccines (NIH)
Asrar Malik (University of Illinois Chicago)
Jalees Rehman (University of Illinois Chicago)
NIH Support: National Heart, Lung, and Blood Institute; National Institute of Allergy and Infectious Diseases
Protein Mapping Study Reveals Valuable Clues for COVID-19 Drug Development
Posted on by Dr. Francis Collins
One way to fight COVID-19 is with drugs that directly target SARS-CoV-2, the novel coronavirus that causes the disease. That’s the strategy employed by remdesivir, the only antiviral drug currently authorized by the U.S. Food and Drug Administration to treat COVID-19. Another promising strategy is drugs that target the proteins within human cells that the virus needs to infect, multiply, and spread.
With the aim of developing such protein-targeted antiviral drugs, a large, international team of researchers, funded in part by the NIH, has precisely and exhaustively mapped all of the interactions that take place between SARS-CoV-2 proteins and the human proteins found within infected host cells. They did the same for the related coronaviruses: SARS-CoV-1, the virus responsible for outbreaks of Severe Acute Respiratory Syndrome (SARS), which ended in 2004; and MERS-CoV, the virus that causes the now-rare Middle East Respiratory Syndrome (MERS).
The goal, as reported in the journal Science, was to use these protein “interactomes” to uncover vulnerabilities shared by all three coronaviruses. The hope is that the newfound knowledge about these shared proteins—and the pathways to which they belong—will inform efforts to develop new kinds of broad-spectrum antiviral therapeutics for use in the current and future coronavirus outbreaks.
Facilitated by the Quantitative Biosciences Institute Research Group, the team, which included David E. Gordon and Nevan Krogan, University of California, San Francisco, and hundreds of other scientists from around the world, successfully mapped nearly 400 protein-protein interactions between SARS-CoV-2 and human proteins.
You can see one of these interactions in the video above. The video starts out with an image of the Orf9b protein of SARS-CoV-2, which normally consists of two linked molecules (blue and orange). But researchers discovered that Orf9b dissociates into a single molecule (orange) when it interacts with the human protein TOM70 (teal). Through detailed structural analysis using cryo-electron microscopy (cryo-EM), the team went on to predict that this interaction may disrupt a key interaction between TOM70 and another human protein called HSP90.
While further study is needed to understand all the details and their implications, it suggests that this interaction may alter important aspects of the human immune response, including blocking interferon signals that are crucial for sounding the alarm to prevent serious illness. While there is no drug immediately available to target Orf9b or TOM70, the findings point to this interaction as a potentially valuable target for treating COVID-19 and other diseases caused by coronaviruses.
This is just one intriguing example out of 389 interactions between SARS-CoV-2 and human proteins uncovered in the new study. The researchers also identified 366 interactions between human and SARS-CoV-1 proteins and 296 for MERS-CoV. They were especially interested in shared interactions that take place between certain human proteins and the corresponding proteins in all three coronaviruses.
To learn more about the significance of these protein-protein interactions, the researchers conducted a series of studies to find out how disrupting each of the human proteins influences SARS-CoV-2’s ability to infect human cells. These studies narrowed the list to 73 human proteins that the virus depends on to replicate.
Among them were the receptor for an inflammatory signaling molecule called IL-17, which has been suggested as an indicator of COVID-19 severity. Two other human proteins—PGES-2 and SIGMAR1—were of particular interest because they are targets of existing drugs, including the anti-inflammatory indomethacin for PGES-2 and antipsychotics like haloperidol for SIGMAR1.
To connect the molecular-level data to existing clinical information for people with COVID-19, the researchers looked to medical billing data for nearly 740,000 Americans treated for COVID-19. They then zeroed in on those individuals who also happened to have been treated with drugs targeting PGES-2 or SIGMAR1. And the results were quite striking.
They found that COVID-19 patients taking indomethacin were less likely than those taking an anti-inflammatory that doesn’t target PGES-2 to require treatment at a hospital. Similarly, COVID-19 patients taking antipsychotic drugs like haloperidol that target SIGMAR1 were half as likely as those taking other types of antipsychotic drugs to require mechanical ventilation.
More research is needed before we can think of testing these or similar drugs against COVID-19 in human clinical trials. Yet these findings provide a remarkable demonstration of how basic molecular and structural biological findings can be combined with clinical data to yield valuable new clues for treating COVID-19 and other viral illnesses, perhaps by repurposing existing drugs. Not only is NIH-supported basic science essential for addressing the challenges of the current pandemic, it is building a strong foundation of fundamental knowledge that will make us better prepared to deal with infectious disease threats in the future.
Reference:
[1] Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Gordon DE et al. Science. 2020 Oct 15:eabe9403.
Links:
Coronavirus (COVID-19) (NIH)
Krogan Lab (University of California, San Francisco)
NIH Support: National Institute of Allergy and Infectious Diseases; National Institute of Neurological Disorders and Stroke; National Institute of General Medical Sciences
Immune T Cells May Offer Lasting Protection Against COVID-19
Posted on by Dr. Francis Collins

Much of the study on the immune response to SARS-CoV-2, the novel coronavirus that causes COVID-19, has focused on the production of antibodies. But, in fact, immune cells known as memory T cells also play an important role in the ability of our immune systems to protect us against many viral infections, including—it now appears—COVID-19.
An intriguing new study of these memory T cells suggests they might protect some people newly infected with SARS-CoV-2 by remembering past encounters with other human coronaviruses. This might potentially explain why some people seem to fend off the virus and may be less susceptible to becoming severely ill with COVID-19.
The findings, reported in the journal Nature, come from the lab of Antonio Bertoletti at the Duke-NUS Medical School in Singapore [1]. Bertoletti is an expert in viral infections, particularly hepatitis B. But, like so many researchers around the world, his team has shifted their focus recently to help fight the COVID-19 pandemic.
Bertoletti’s team recognized that many factors could help to explain how a single virus can cause respiratory, circulatory, and other symptoms that vary widely in their nature and severity—as we’ve witnessed in this pandemic. One of those potential factors is prior immunity to other, closely related viruses.
SARS-CoV-2 belongs to a large family of coronaviruses, six of which were previously known to infect humans. Four of them are responsible for the common cold. The other two are more dangerous: SARS-CoV-1, the virus responsible for the outbreak of Severe Acute Respiratory Syndrome (SARS), which ended in 2004; and MERS-CoV, the virus that causes Middle East Respiratory Syndrome (MERS), first identified in Saudi Arabia in 2012.
All six previously known coronaviruses spark production of both antibodies and memory T cells. In addition, studies of immunity to SARS-CoV-1 have shown that T cells stick around for many years longer than acquired antibodies. So, Bertoletti’s team set out to gain a better understanding of T cell immunity against the novel coronavirus.
The researchers gathered blood samples from 36 people who’d recently recovered from mild to severe COVID-19. They focused their attention on T cells (including CD4 helper and CD8 cytotoxic, both of which can function as memory T cells). They identified T cells that respond to the SARS-CoV-2 nucleocapsid, which is a structural protein inside the virus. They also detected T cell responses to two non-structural proteins that SARS-CoV-2 needs to make additional copies of its genome and spread. The team found that all those recently recovered from COVID-19 produced T cells that recognize multiple parts of SARS-CoV-2.
Next, they looked at blood samples from 23 people who’d survived SARS. Their studies showed that those individuals still had lasting memory T cells today, 17 years after the outbreak. Those memory T cells, acquired in response to SARS-CoV-1, also recognized parts of SARS-CoV-2.
Finally, Bertoletti’s team looked for such T cells in blood samples from 37 healthy individuals with no history of either COVID-19 or SARS. To their surprise, more than half had T cells that recognize one or more of the SARS-CoV-2 proteins under study here. It’s still not clear if this acquired immunity stems from previous infection with coronaviruses that cause the common cold or perhaps from exposure to other as-yet unknown coronaviruses.
What’s clear from this study is our past experiences with coronavirus infections may have something important to tell us about COVID-19. Bertoletti’s team and others are pursuing this intriguing lead to see where it will lead—not only in explaining our varied responses to the virus, but also in designing new treatments and optimized vaccines.
Reference:
[1] SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Le Bert N, Tan AT, Kunasegaran K, et al. Nature. 2020 July 15. [published online ahead of print]
Links:
Coronavirus (COVID-19) (NIH)
Overview of the Immune System (National Institute of Allergy and Infectious Diseases/NIAID)
Bertoletti Lab (Duke-NUS Medical School, Singapore)
Promising Treatment for New Human Coronavirus
Posted on by Dr. Francis Collins

Caption: Transmission electron micrograph of novel coronavirus
Credit: Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, NIH
In Fall 2012 a new coronavirus appeared on the global public health radar. The virus has caused 17 cases of severe respiratory disease in the Middle East and Europe, and 11 of these people died. This new virus attracted immediate attention because of the high fatality rate—and because it was in the same family as the virus that caused the global outbreak of severe acute respiratory syndrome (SARS) in 2003, which sickened more than 8,000 people.