Adding Letters to the DNA Alphabet
Posted on by Dr. Francis Collins
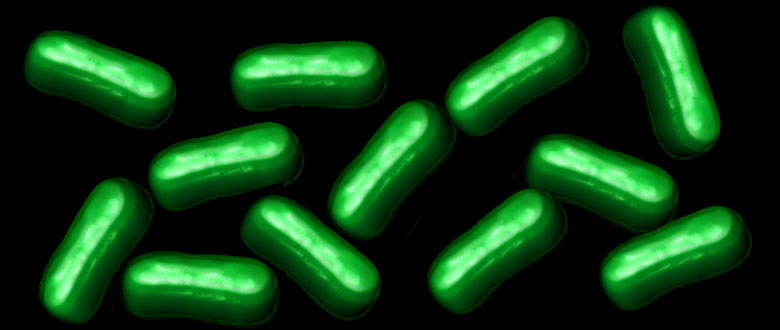
Credit: William B. Kiosses
The recipes for life, going back billions of years to the earliest single-celled organisms, are encoded in a DNA alphabet of just four letters. But is four as high as the DNA code can go? Or, as researchers have long wondered, is it chemically and biologically possible to expand the DNA code by a couple of letters?
A team of NIH-funded researchers is now answering these provocative questions. The researchers recently engineered a semi-synthetic bacterium containing DNA with six letters, including two extra nucleotides [1, 2]. Now, in a report published in Nature, they’ve taken the next critical step [3]. They show that bacteria, like those in the photo, are not only capable of reliably passing on to the next generation a DNA code of six letters, they can use that expanded genetic information to produce novel proteins unlike any found in nature.
As a quick refresher, DNA molecules form a double helix, which looks much like a twisted ladder. Each rung of the ladder contains two nucleotides bases—the all-important As, Gs, Ts, and Cs. Those bases always pair up in a particular way, with A pairing with T and C with G. That pairing is essential for DNA’s ability to unwind and make faithful copies of itself before cells divide.
When a team led by Floyd Romesberg at The Scripps Research Institute, La Jolla, CA, first set out to expand the DNA code, the first challenge was a chemical one: to find two new nucleotide letters that would pair up like A-T and G-C. It took about a dozen years, but after studying in test tubes a long list of chemical candidates in thousands of combinations, they finally added a new rung to the ladder. They paired up two promising new nucleotide bases, dubbed X and Y. (At the bottom of this blog, you can see the actual structures.)
The next challenge was to find out whether X and Y could be integrated into the DNA of a living organism. The researchers looked to a harmless strain of the bacterium Escherichia coli, which is commonly studied in the lab. It took plenty of tinkering, but eventually they succeeded. They inserted the new X-Y base pair into the bacteria’s DNA. They then showed that the single-celled organism could reliably copy the expanded DNA code and pass it on to the next generation.
To make it work, they engineered the E. coli in a lab dish to produce a special transporter protein normally found in algae. The special protein allowed the engineered bacteria to take up the biochemical ingredients needed to produce the two unnatural nucleotides. As long as those key ingredients were added, the bacteria survived and could reproduce using that expanded DNA code.
But could those bacteria also retrieve information from DNA containing those two new letters? First, another refresher: The sequence of nucleotide bases running along each strand of DNA, each half of the ladder, form a special language consisting of three-letter “words,” or codons. Each codon within protein-coding gene sequences codes for one of 20 amino acids, the building blocks of proteins.
For cells to produce proteins from gene sequences, DNA is first transcribed into RNA messages. Those messages get translated into proteins. It happens as molecules called transfer RNAs recognize the three-letter codons and ferry in the right amino acids. So, the question was: could the bacteria transcribe messages including codons with all six letters and then use those messages to produce brand new kinds of proteins?
To find out, the researchers inserted into the E. coli cells a gene sequence containing the six-letter code along with a corresponding transfer RNA. That transfer RNA was equipped to recognize a new X or Y-containing codon and bring in an amino acid not found in nature.
To let them know whether the experiment was working, the researchers incorporated those new letters into a gene encoding an easy to see green fluorescent protein (GFP). And, it worked. The bacteria efficiently decoded the unnatural codon. As a result, the bacteria started glowing with green fluorescent proteins containing the unnatural amino acid, just as you can see in the picture above.
An extra two letters might not sound like a lot, but it now vastly expands the number of possible three-letter “words” that can be written in DNA. In fact, the expanded code boosts the number of possible codons and corresponding amino acids from 20 to 172!
Of course, caution is warranted. It’s worth reiterating that the engineered E. coli can’t survive without being fed the right ingredients to make the unnatural gene sequences and the proteins they encode. They’d quickly die if they were to somehow escape from the lab. Ethical concerns aside, these semi-synthetic E. coli have potential for producing all kinds of interesting new proteins and perhaps other materials as well.
An exciting potential application is in protein therapeutics, or engineering proteins in the lab to treat disease. In the last 25 years, protein therapeutic products, such as insulin and human growth hormone, have become more common in medicine [4]. But those proteins have been limited to the 20 amino acids found in nature.
With this latest advance, it’s now possible to explore proteins with interesting new structures and functions. Romesberg says they are already pursuing a couple of the most obvious applications, including modifying existing or promising engineered proteins in ways that could make them more effective.
They also continue to work on the basics, such as showing it’s possible to produce proteins that include multiple unnatural amino acids and on a scale that would be useful for producing new therapeutics. I can’t wait to see where it leads.
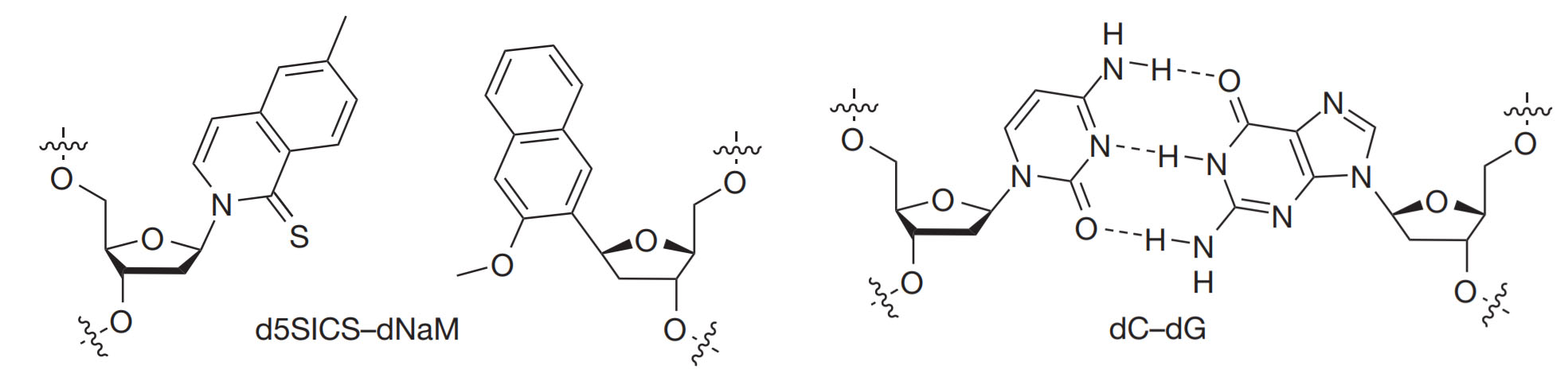
For the experts: On the left you can see the actual chemical structures of X (dNaM) and Y (dTPT3) used by Romesberg and colleagues. On the right is shown the natural base pairing of dA and dT, characteristic of all living species.
Source: Adapted from Nature. 2014 May 15;509(7500):385-388.
References:
[1] A semi-synthetic organism with an expanded genetic alphabet. Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Corrêa IR Jr, Romesberg FE. Nature. 2014 May 15;509(7500):385-8.
[2] A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Zhang Y, Lamb BM, Feldman AW, Zhou AX, Lavergne T, Li L, Romesberg FE. Proc Natl Acad Sci U S A. 2017 Feb 7;114(6):1317-1322.
[3] A semi-synthetic organism that stores and retrieves increased genetic information. Zhang Y, Ptacin JL, Fischer EC, Aerni HR, Caffaro CE, San Jose K, Feldman AW, Turner CR, Romesberg FE. Nature. 2017 Nov 29;551(7682):644-647.
[4] Protein therapeutics: a summary and pharmacological classification. Leader B, Baca QJ, Golan DE. Nat Rev Drug Discov. 2008 Jan;7(1):21
Links:
DNA (National Human Genome Research Institute/NIH)
Romesberg Lab (The Scripps Research Institute, La Jolla, CA)
NIH Support: National Institute of General Medical Sciences
Share this:
- Click to share on LinkedIn (Opens in new window)
- Click to share on Pinterest (Opens in new window)
- Click to share on Tumblr (Opens in new window)
- Click to share on Reddit (Opens in new window)
- Click to share on Telegram (Opens in new window)
- Click to share on WhatsApp (Opens in new window)
- Click to print (Opens in new window)
Posted In: Health, Science, technology
Tags: amino acids, bacteria, bioengineering, codon, DNA, DNA alphabet, DNA code, E. coli, expanded DNA code, genetic code, green fluorescent protein, nucleotides, protein, protein therapeutics, transfer RNA

this is huge!
Perfect article. Thanks for sharing
thank you for sharing this article