cancer
An Evolutionary Guide to New Immunotherapies
Posted on by Dr. Francis Collins

One of the best ways to learn how something works is to understand how it’s built. How it came to be. That’s true not only if you play a guitar or repair motorcycle engines, but also if you study the biological systems that make life possible. Evolutionary studies, comparing the development of these systems across animals and organisms, are now leading to many unexpected biological discoveries and promising possibilities for preventing and treating human disease.
While there are many evolutionary questions to ask, Brenda Bass, a distinguished biochemist at University of Utah, Salt Lake City, has set her sights on a particularly profound one: How has innate immunity evolved through the millennia in all living things, including humans? Innate immunity is the immune system’s frontline defense, the first responders that take control of an emerging infectious situation and, if needed, signal for backup.
Exploring the millennia for clues about innate immunity takes a special team, and Bass has assembled a talented one. It includes her Utah colleague Nels Elde, a geneticist; immunologist Dan Stetson, University of Washington, Seattle; and biochemist Jane Jackman, Ohio State University, Columbus.
With a 2020 NIH Director’s Transformative Research Award, this hard-working team will embark on studies looking back at 450 million years of evolution: the point in time when animals diverged to develop very distinct methods of innate immune defense [1]. The team members hope to uncover new possibilities encoded in the innate immune system, especially those that might be latent but still workable. The researchers will then explore whether their finds can be repurposed not only to boost our body’s natural response to external threats but also to internal threats like cancer.
Bass brings a unique perspective to the project. As a postdoc in the 1980s, she stumbled upon a whole new class of enzymes, called ADARs, that edit RNA [2]. Their function was mysterious at the time. It turns out that ADARs specifically edit a molecule called double-stranded RNA (dsRNA). When viruses infect cells in animals, including humans, they make dsRNA, which the innate immune system detects as a sign that a cell has been invaded.
It also turns out that animal cells make their own dsRNA. Over the years, Bass and her lab have identified thousands of dsRNAs made in animal cells—in fact, a significant number of human genes produce dsRNA [3]. Also interesting, ADARs are crucial to marking our own dsRNA as “self” to avoid triggering an immune response when we don’t need it [4].
Bass and others have found that evolution has produced dramatic differences in the biochemical pathways powering the innate immune system. In vertebrate animals, dsRNA leads to release of the immune chemical interferon, a signaling pathway that invertebrate species don’t have. Instead, in response to detecting dsRNA from an invader, and repelling it, worms and other invertebrates trigger a gene-silencing pathway known as RNA interference, or RNAi.
With the new funding, Bass and team plan to mix and match immune strategies from simple and advanced species, across evolutionary time, to craft an entirely new set of immune tools to fight disease. The team will also build new types of targeted immunotherapies based on the principles of innate immunity. Current immunotherapies, which harness a person’s own immune system to fight disease, target infections, autoimmune disorders, and cancer. But they work through our second-line adaptive immune response, which is a biological system unique to vertebrates.
Bass and her team will first hunt for more molecules like ADARs: innate immune checkpoints, as they refer to them. The name comes from a functional resemblance to the better-known adaptive immune checkpoints PD-1 and CTLA-4, which sparked a revolution in cancer immunotherapy. The team will run several screens that sort molecules successful at activating innate immune responses—both in invertebrates and in mammals—hoping to identify a range of durable new immune switches that evolution skipped over but that might be repurposed today.
Another intriguing direction for this research stems from the observation that decreasing normal levels of ADARs in tumors kickstarts innate immune responses that kill cancer cells [5]. Along these lines, the scientists plan to test newly identified immune switches to look for novel ways to fight cancer where existing approaches have not worked.
Evolution is the founding principle for all of biology—organisms learn from what works to improve their ability to survive. In this case, research to re-examine such lessons and apply them for new uses may help transform bygone evolution into a therapeutic revolution!
References:
[1] Evolution of adaptive immunity from transposable elements combined with innate immune systems. Koonin EV, Krupovic M. Nat Rev Genet. 2015 Mar;16(3):184-192.
[2] A developmentally regulated activity that unwinds RNA duplexes. Bass BL, Weintraub H. Cell. 1987 Feb 27;48(4):607-613.
[3] Mapping the dsRNA World. Reich DP, Bass BL. Cold Spring Harb Perspect Biol. 2019 Mar 1;11(3):a035352.
[4] To protect and modify double-stranded RNA – the critical roles of ADARs in development, immunity and oncogenesis. Erdmann EA, Mahapatra A, Mukherjee P, Yang B, Hundley HA. Crit Rev Biochem Mol Biol. 2021 Feb;56(1):54-87.
[5] Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, et al. Nature. 2019 Jan;565(7737):43-48.
Links:
Bass Lab (University of Utah, Salt Lake City)
Elde Lab (University of Utah)
Jackman Lab (Ohio State University, Columbus)
Stetson Lab (University of Washington, Seattle)
Bass/Elde/Jackman/Stetson Project Information (NIH RePORTER)
NIH Director’s Transformative Research Award Program (Common Fund)
NIH Support: Common Fund; National Cancer Institute
Welcoming First Lady Jill Biden to NIH!
Posted on by Dr. Francis Collins

It was wonderful to have First Lady Jill Biden pay a virtual visit to NIH on February 3, 2021, on the eve of World Cancer Day. Dr. Biden joined me, National Cancer Institute (NCI) Director Ned Sharpless, and several NCI scientists to discuss recent advances in fighting cancer. On behalf of the entire NIH community, I thanked the First Lady for her decades of advocacy on behalf of cancer education, prevention, and research. To view the event, go to 53:20 in this video. Credit: Adapted from White House video.
The People’s Picks for Best Posts
Posted on by Dr. Francis Collins
It’s 2021—Happy New Year! Time sure flies in the blogosphere. It seems like just yesterday that I started the NIH Director’s Blog to highlight recent advances in biology and medicine, many supported by NIH. Yet it turns out that more than eight years have passed since this blog got rolling and we are fast approaching my 1,000th post!
I’m pleased that millions of you have clicked on these posts to check out some very cool science and learn more about NIH and its mission. Thanks to the wonders of social media software, we’ve been able to tally up those views to determine each year’s most-popular post. So, I thought it would be fun to ring in the New Year by looking back at a few of your favorites, sort of a geeky version of a top 10 countdown or the People’s Choice Awards. It was interesting to see what topics generated the greatest interest. Spoiler alert: diet and exercise seemed to matter a lot! So, without further ado, I present the winners:
2013: Fighting Obesity: New Hopes from Brown Fat. Brown fat, one of several types of fat made by our bodies, was long thought to produce body heat rather than store energy. But Shingo Kajimura and his team at the University of California, San Francisco, showed in a study published in the journal Nature, that brown fat does more than that. They discovered a gene that acts as a molecular switch to produce brown fat, then linked mutations in this gene to obesity in humans.
What was also nice about this blog post is that it appeared just after Kajimura had started his own lab. In fact, this was one of the lab’s first publications. One of my goals when starting the blog was to feature young researchers, and this work certainly deserved the attention it got from blog readers. Since highlighting this work, research on brown fat has continued to progress, with new evidence in humans suggesting that brown fat is an effective target to improve glucose homeostasis.
2014: In Memory of Sam Berns. I wrote this blog post as a tribute to someone who will always be very near and dear to me. Sam Berns was born with Hutchinson-Gilford progeria syndrome, one of the rarest of rare diseases. After receiving the sad news that this brave young man had passed away, I wrote: “Sam may have only lived 17 years, but in his short life he taught the rest of us a lot about how to live.”
Affecting approximately 400 people worldwide, progeria causes premature aging. Without treatment, children with progeria, who have completely normal intellectual development, die of atherosclerotic cardiovascular disease, on average in their early teens.
From interactions with Sam and his parents in the early 2000s, I started to study progeria in my NIH lab, eventually identifying the gene responsible for the disorder. My group and others have learned a lot since then. So, it was heartening last November when the Food and Drug Administration approved the first treatment for progeria. It’s an oral medication called Zokinvy (lonafarnib) that helps prevent the buildup of defective protein that has deadly consequences. In clinical trials, the drug increased the average survival time of those with progeria by more than two years. It’s a good beginning, but we have much more work to do in the memory of Sam and to help others with progeria. Watch for more about new developments in applying gene editing to progeria in the next few days.
2015: Cytotoxic T Cells on Patrol. Readers absolutely loved this post. When the American Society of Cell Biology held its first annual video competition, called CellDance, my blog featured some of the winners. Among them was this captivating video from Alex Ritter, then working with cell biologist Jennifer Lippincott-Schwartz of NIH’s Eunice Kennedy Shriver National Institute of Child Health and Human Development. The video stars a roving, specialized component of our immune system called cytotoxic T cells. Their job is to seek out and destroy any foreign or detrimental cells. Here, these T cells literally convince a problem cell to commit suicide, a process that takes about 10 minutes from detection to death.
These cytotoxic T cells are critical players in cancer immunotherapy, in which a patient’s own immune system is enlisted to control and, in some cases, even cure the cancer. Cancer immunotherapy remains a promising area of research that continues to progress, with a lot of attention now being focused on developing immunotherapies for common, solid tumors like breast cancer. Ritter is currently completing a postdoctoral fellowship in the laboratory of Ira Mellman, Genentech, South San Francisco. His focus has shifted to how cancer cells protect themselves from T cells. And video buffs—get this—Ritter says he’s now created even cooler videos that than the one in this post.
2016: Exercise Releases Brain-Healthy Protein. The research literature is pretty clear: exercise is good for the brain. In this very popular post, researchers led by Hyo Youl Moon and Henriette van Praag of NIH’s National Institute on Aging identified a protein secreted by skeletal muscle cells to help explore the muscle-brain connection. In a study in Cell Metabolism, Moon and his team showed that this protein called cathepsin B makes its way into the brain and after a good workout influences the development of new neural connections. This post is also memorable to me for the photo collage that accompanied the original post. Why? If you look closely at the bottom right, you’ll see me exercising—part of my regular morning routine!
2017: Muscle Enzyme Explains Weight Gain in Middle Age. The struggle to maintain a healthy weight is a lifelong challenge for many of us. While several risk factors for weight gain, such as counting calories, are within our control, there’s a major one that isn’t: age. Jay Chung, a researcher with NIH’s National Heart, Lung, and Blood Institute, and his team discovered that the normal aging process causes levels of an enzyme called DNA-PK to rise in animals as they approach middle age. While the enzyme is known for its role in DNA repair, their studies showed it also slows down metabolism, making it more difficult to burn fat.
Since publishing this paper in Cell Metabolism, Chung has been busy trying to understand how aging increases the activity of DNA-PK and its ability to suppress renewal of the cell’s energy-producing mitochondria. Without renewal of damaged mitochondria, excess oxidants accumulate in cells that then activate DNA-PK, which contributed to the damage in the first place. Chung calls it a “vicious cycle” of aging and one that we’ll be learning more about in the future.
2018: Has an Alternative to Table Sugar Contributed to the C. Diff. Epidemic? This impressive bit of microbial detective work had blog readers clicking and commenting for several weeks. So, it’s no surprise that it was the runaway People’s Choice of 2018.
Clostridium difficile (C. diff) is a common bacterium that lives harmlessly in the gut of most people. But taking antibiotics can upset the normal balance of healthy gut microbes, allowing C. diff. to multiply and produce toxins that cause inflammation and diarrhea.
In the 2000s, C. diff. infections became far more serious and common in American hospitals, and Robert Britton, a researcher at Baylor College of Medicine, Houston, wanted to know why. He and his team discovered that two subtypes of C. diff have adapted to feed on the sugar trehalose, which was approved as a food additive in the United States during the early 2000s. The team’s findings, published in the journal Nature, suggested that hospitals and nursing homes battling C. diff. outbreaks may want to take a closer look at the effect of trehalose in the diet of their patients.
2019: Study Finds No Benefit for Dietary Supplements. This post that was another one that sparked a firestorm of comments from readers. A team of NIH-supported researchers, led by Fang Fang Zhang, Tufts University, Boston, found that people who reported taking dietary supplements had about the same risk of dying as those who got their nutrients through food. What’s more, the mortality benefits associated with adequate intake of vitamin A, vitamin K, magnesium, zinc, and copper were limited to amounts that are available from food consumption. The researchers based their conclusion on an analysis of the well-known National Health and Nutrition Examination Survey (NHANES) between 1999-2000 and 2009-2010 survey data. The team, which reported its data in the Annals of Internal Medicine, also uncovered some evidence suggesting that certain supplements might even be harmful to health when taken in excess.
2020: Genes, Blood Type Tied to Risk of Severe COVID-19. Typically, my blog focuses on research involving many different diseases. That changed in 2020 due to the emergence of a formidable public health challenge: the coronavirus disease 2019 (COVID-19) pandemic. Since last March, the blog has featured 85 posts on COVID-19, covering all aspects of the research response and attracting more visitors than ever. And which post got the most views? It was one that highlighted a study, published last June in the New England Journal of Medicine, that suggested the clues to people’s variable responses to COVID-19 may be found in our genes and our blood types.
The researchers found that gene variants in two regions of the human genome are associated with severe COVID-19 and correspondingly carry a greater risk of COVID-19-related death. The two stretches of DNA implicated as harboring risks for severe COVID-19 are known to carry some intriguing genes, including one that determines blood type and others that play various roles in the immune system.
In fact, the findings suggest that people with blood type A face a 50 percent greater risk of needing oxygen support or a ventilator should they become infected with the novel coronavirus. In contrast, people with blood type O appear to have about a 50 percent reduced risk of severe COVID-19.
That’s it for the blog’s year-by-year Top Hits. But wait! I’d also like to give shout outs to the People’s Choice winners in two other important categories—history and cool science images.
Top History Post: HeLa Cells: A New Chapter in An Enduring Story. Published in August 2013, this post remains one of the blog’s greatest hits with readers. The post highlights science’s use of cancer cells taken in the 1950s from a young Black woman named Henrietta Lacks. These “HeLa” cells had an amazing property not seen before: they could be grown continuously in laboratory conditions. The “new chapter” featured in this post is an agreement with the Lacks family that gives researchers access to the HeLa genome data, while still protecting the family’s privacy and recognizing their enormous contribution to medical research. And the acknowledgments rightfully keep coming from those who know this remarkable story, which has been chronicled in both book and film. Recently, the U.S. Senate and House of Representatives passed the Henrietta Lacks Enhancing Cancer Research Act to honor her extraordinary life and examine access to government-funded cancer clinical trials for traditionally underrepresented groups.
Top Snapshots of Life: A Close-up of COVID-19 in Lung Cells. My blog posts come in several categories. One that you may have noticed is “Snapshots of Life,” which provides a showcase for cool images that appear in scientific journals and often dominate Science as Art contests. My blog has published dozens of these eye-catching images, representing a broad spectrum of the biomedical sciences. But the blog People’s Choice goes to a very recent addition that reveals exactly what happens to cells in the human airway when they are infected with the coronavirus responsible for COVID-19. This vivid image, published in the New England Journal of Medicine, comes from the lab of pediatric pulmonologist Camille Ehre, University of North Carolina at Chapel Hill. This image squeezed in just ahead of another highly popular post from Steve Ramirez, Boston University, in 2019 that showed “What a Memory Looks Like.”
As we look ahead to 2021, I want to thank each of my blog’s readers for your views and comments over the last eight years. I love to hear from you, so keep on clicking! I’m confident that 2021 will generate a lot more amazing and bloggable science, including even more progress toward ending the COVID-19 pandemic that made our past year so very challenging.
All of Us: Partnering Together for the Future of Precision Medicine
Posted on by Dr. Francis Collins

Over the past year, it’s been so inspiring to watch tens of thousands of people across the country selflessly step forward for vaccine trials and other research studies to combat COVID-19. And they are not alone. Many generous folks are volunteering to take part in other types of NIH-funded research that will improve health all across the spectrum, including the more than 360,000 who’ve already enrolled in the pioneering All of Us Research Program.
Now in its second year, All of Us is building a research community of 1 million participant partners to help us learn more about how genetics, environment, and lifestyle interact to influence disease and affect health. So far, more than 80 percent of participants who have completed all the initial enrollment steps are Black, Latino, rural, or from other communities historically underrepresented in biomedical research.
This community will build a diverse foundation for precision medicine, in which care is tailored to the individual, not the average patient as is now often the case. What’s also paradigm shifting about All of Us is its core value of sharing information back with participants about themselves. It is all done responsibly through each participant’s personal All of Us online account and with an emphasis on protecting privacy.
All of Us participants share their health information in many ways, such as taking part in surveys, offering access to their electronic health records, and providing biosamples (blood, urine, and/or saliva). In fact, researchers recently began genotyping and sequencing the DNA in some of those biosamples, and then returning results from analyses to participants who’ve indicated they’d like to receive such information. This first phase of genotyping DNA analysis will provide insights into their genetic ancestry and four traits, including bitter taste perception and tolerance for lactose.
Results of a second sequencing phase of DNA analysis will likely be ready in the coming year. These personalized reports will give interested participants information about how their bodies are likely to react to certain medications and about whether they face an increased risk of developing certain health conditions, such as some types of cancer or heart disease. To help participants better understand the results, they can make a phone appointment with a genetic counselor who is affiliated with the program.
This week, I had the pleasure of delivering the keynote address at the All of Us Virtual Face-to-Face. This lively meeting was attended by a consortium of more than 2,000 All of Us senior staff, program leads with participating healthcare provider organizations and federally qualified health centers, All of Us-supported researchers, community partners, and the all-important participant ambassadors.
If you are interested in becoming part of the All of Us community, I welcome you—there’s plenty of time to get involved! To learn more, just go to Join All of Us.
Links:
All of Us Research Program (NIH)
Join All of Us (NIH)
Discussing Cancer Health Disparities
Posted on by Dr. Francis Collins

See the Human Cardiovascular System in a Whole New Way
Posted on by Dr. Francis Collins
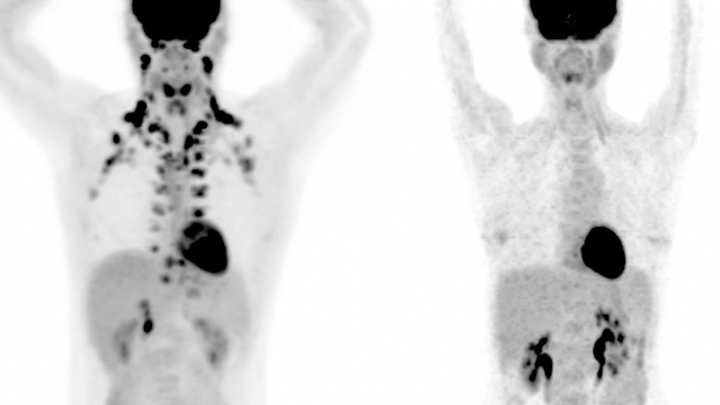
Watch this brief video and you might guess you’re seeing an animated line drawing, gradually revealing a delicate take on a familiar system: the internal structures of the human body. But this movie doesn’t capture the work of a talented sketch artist. It was created using the first 3D, full-body imaging device using positron emission tomography (PET).
The device is called an EXPLORER (EXtreme Performance LOng axial REsearch scanneR) total-body PET scanner. By pairing this scanner with an advanced method for reconstructing images from vast quantities of data, the researchers can make movies.
For this movie in particular, the researchers injected small amounts of a short-lived radioactive tracer—an essential component of all PET scans—into the lower leg of a study volunteer. They then sat back as the scanner captured images of the tracer moving up the leg and into the body, where it enters the heart. The tracer moves through the heart’s right ventricle to the lungs, back through the left ventricle, and up to the brain. Keep watching, and, near the 30-second mark, you will see in closer focus a haunting capture of the beating heart.
This groundbreaking scanner was developed and tested by Jinyi Qi, Simon Cherry, Ramsey Badawi, and their colleagues at the University of California, Davis [1]. As the NIH-funded researchers reported recently in Proceedings of the National Academy of Sciences, their new scanner can capture dynamic changes in the body that take place in a tenth of a second [2]. That’s faster than the blink of an eye!
This movie is composed of frames captured at 0.1-second intervals. It highlights a feature that makes this scanner so unique: its ability to visualize the whole body at once. Other medical imaging methods, including MRI, CT, and traditional PET scans, can be used to capture beautiful images of the heart or the brain, for example. But they can’t show what’s happening in the heart and brain at the same time.
The ability to capture the dynamics of radioactive tracers in multiple organs at once opens a new window into human biology. For example, the EXPLORER system makes it possible to measure inflammation that occurs in many parts of the body after a heart attack, as well as to study interactions between the brain and gut in Parkinson’s disease and other disorders.
EXPLORER also offers other advantages. It’s extra sensitive, which enables it to capture images other scanners would miss—and with a lower dose of radiation. It’s also much faster than a regular PET scanner, making it especially useful for imaging wiggly kids. And it expands the realm of research possibilities for PET imaging studies. For instance, researchers might repeatedly image a person with arthritis over time to observe changes that may be related to treatments or exercise.
Currently, the UC Davis team is working with colleagues at the University of California, San Francisco to use EXPLORER to enhance our understanding of HIV infection. Their preliminary findings show that the scanner makes it easier to capture where the human immunodeficiency virus (HIV), the cause of AIDS, is lurking in the body by picking up on signals too weak to be seen on traditional PET scans.
While the research potential for this scanner is clearly vast, it also holds promise for clinical use. In fact, a commercial version of the scanner, called uEXPLORER, has been approved by the FDA and is in use at UC Davis [3]. The researchers have found that its improved sensitivity makes it much easier to detect cancers in patients who are obese and, therefore, harder to image well using traditional PET scanners.
As soon as the COVID-19 outbreak subsides enough to allow clinical research to resume, the researchers say they’ll begin recruiting patients with cancer into a clinical study designed to compare traditional PET and EXPLORER scans directly.
As these researchers, and other researchers around the world, begin to put this new scanner to use, we can look forward to seeing many more remarkable movies like this one. Imagine what they will reveal!
References:
[1] First human imaging studies with the EXPLORER total-body PET scanner. Badawi RD, Shi H, Hu P, Chen S, Xu T, Price PM, Ding Y, Spencer BA, Nardo L, Liu W, Bao J, Jones T, Li H, Cherry SR. J Nucl Med. 2019 Mar;60(3):299-303.
[2] Subsecond total-body imaging using ultrasensitive positron emission tomography. Zhang X, Cherry SR, Xie Z, Shi H, Badawi RD, Qi J. Proc Natl Acad Sci U S A. 2020 Feb 4;117(5):2265-2267.
[3] “United Imaging Healthcare uEXPLORER Total-body Scanner Cleared by FDA, Available in U.S. Early 2019.” Cision PR Newswire. January 22, 2019.
Links:
Positron Emission Tomography (PET) (NIH Clinical Center)
EXPLORER Total-Body PET Scanner (University of California, Davis)
Cherry Lab (UC Davis)
Badawi Lab (UC Davis Medical Center, Sacramento)
NIH Support: National Cancer Institute; National Institute of Biomedical Imaging and Bioengineering; Common Fund
Prostate Cancer: Combined Biopsy Strategy Makes for More Accurate Diagnosis
Posted on by Dr. Francis Collins

Last year, nearly 175,000 American men were diagnosed with prostate cancer [1]. Most got the bad news after a blood test or physical exam raised concerns that warranted a biopsy of the prostate, a walnut-sized gland just below the bladder.
Traditional biopsies sample tissue from 12 systematically placed points within the prostate that are blind to tumor locations. Such procedures have helped to save many lives, but are prone to missing or misclassifying prostate cancers, which has led doctors both to over and under treat their patients.
Now, there may be a better approach. In a study of more than 2,000 men, NIH researchers and their colleagues recently found that combining the 12-point biopsy with magnetic resonance imaging (MRI)-targeted biopsy during the same session more accurately diagnoses prostate cancer than either technique alone [2].
The findings address a long-standing challenge in prostate cancer diagnostics: performing a thorough prostate biopsy to allow a pathologist to characterize as accurately as possible the behavior of a tumor. Some prostate tumors are small, slow growing, and can be monitored closely without treatment. Other tumors are aggressive and can grow rapidly, requiring immediate intervention with hormonal therapy, radiation, or surgery.
But performing a thorough prostate biopsy can run into technical difficulties. The 12-point biopsy blindly samples tissue from across the prostate gland, but it can miss a cancer by not probing in the right places.
Several years ago, researchers at the NIH Clinical Center, Bethesda, MD, envisioned a solution. They’d use specially designed MRI images of a man’s prostate to guide the biopsy needle to areas in the prostate that look suspicious and deserve a closer look under a microscope.
Through a cooperative research-and-development agreement, NIH and the now- Florida-based Philips Healthcare created an office-based, outpatient prostate biopsy device, called UroNav, that was later approved by the Food and Drug Administration. The UroNav system relies on software that overlays MRI images highlighting suspicious areas onto real-time ultrasound images of the prostate that are traditionally used to guide biopsy procedures.
The new technology led to a large clinical study led by Peter Pinto, a researcher with NIH’s National Cancer Institute. The study results, published in 2015, found that the MRI-targeted approach was indeed superior to the 12-point biopsy at detecting aggressive prostate cancers [3].
But some doctors had questions about how best to implement the UroNav system and whether it could replace the 12-point biopsy. The uncertainty led to a second clinical study to nail down more answers, and the results were just published in The New England Journal of Medicine.
The research team enrolled 2,103 men who had visible prostate abnormalities on an MRI. Once in the study, each man underwent both the 12-point blind biopsy and the MRI-targeted approach—all in a single office visit. Based on this two-step approach, 1,312 people were diagnosed with prostate cancer. Of that total, 404 men had evidence of aggressive cancer, and had their prostates surgically removed.
The researchers then compared the diagnoses from each approach alone versus the two combined. The data showed that the combined biopsy found 208 cancers that the standard 12-point biopsy alone would have missed. Adding the MRI-targeted biopsy also helped doctors find and sample the more aggressive cancers. This allowed them to upgrade the diagnosis of 458 cancers to aggressive and in need of more full treatment.
Combining the two approaches also led to more accurate diagnoses. By carefully analyzing the 404 removed prostates and comparing them to the biopsy results, the researchers found the 12-point biopsy missed the most aggressive cancers about 40 percent of the time. But the MRI-targeted approach alone missed it about 30 percent of the time. Combined, they did much better, underestimating the severity of less than 15 percent of the cancers.
Even better, the combined biopsy missed only 3.5 percent of the most aggressive tumors. That’s compared to misses of about 17 percent for the most-aggressive cancers for the 12-point biopsy alone and about 9 percent for MRI-targeted biopsy alone.
It may take time for doctors to change how they detect prostate cancer in their practices. But the findings show that combining both approaches will significantly improve the accuracy of diagnosing prostate cancer. This will, in turn, help to reduce risk of suboptimal treatment (too much or too little) by allowing doctors and patients to feel more confident in the biopsy results. That should come as good news now and in the future for the families and friends of men who will need an accurate prostate biopsy to make informed treatment decisions.
References:
[1] Cancer State Facts: Prostate Cancer. National Cancer Institute Surveillance, Epidemiology, and End Results Program.
[2] MRI-targeted, systematic, and combined biopsy for prostate cancer diagnosis. Ahdoot M, Wilbur AR, Reese SE, Lebastchi AH, Mehralivand S, Gomella PT, Bloom J, Gurram S, Siddiqui M, Pinsky P, Parnes H, Linehan WM, Merino M, Choyke PL, Shih JH, Turkbey B, Wood BJ, Pinto PA. N Engl J Med. 2020 Mar 5;382(10):917-928.
[3] Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. Siddiqui M, Rais-Bahrami, George AK, Rothwax J, Shakir N, Okoro C, Raskolnikov D, Parnes HL, Linehan WM, Merino MJ, Simon RM, Choyke PL, Wood BJ, and Pinto PA. JAMA. 2015 January 27;313(4):390-397.
Links:
Prostate Cancer (National Cancer Institute/NIH)
Video: MRI-Targeted Prostate Biopsy (YouTube)
Pinto Lab (National Cancer Institute/NIH)
NIH Support: National Cancer Institute; NIH Clinical Center
Previous Page Next Page